Abstract
Styrax wuyuanensis S. M. Hwang is an endemic species distributed in China. In this study, we characterized its complete chloroplast genome. The circular genome of S. wuyuanensis is 157,969 bp in length, and includes two inverted repeat (IRa and IRb) regions of 25,954 bp in length separated by a large single copy (LSC) region of 87,575 bp and a small single copy (SSC) region of 18,486 bp. The total GC content of the S. wuyuanensis chloroplast genome is 37.0%, and a total of 132 functional genes are encoded, including 87 protein-coding genes, 37 tRNA, and eight rRNA. The phylogenetic analysis has shown that S. wuyuanensis is positioned in the Styracaceae clade, as a sister taxon to S. faberi and S. fortunei, confirming the close relationship of S. wuyuanensis with the latter two species.
Styrax wuyuanensis S. M. Hwang is a deciduous small shrub only known from the border region of Jiangxi and Anhui Provinces in China, and the type of this species is located in Wuyuan County, Jiangxi Province (Hwang Citation1980). This species can be easily distinguished from all other members of Styrax ser. Cyrta by its subglabrous calyx and pedicel (Li and Fritsch Citation2018). To date, no molecular data of the species have been reported.
In this study, we obtained and reported the complete chloroplast genome sequence of Styrax wuyuanensis (GenBank accession number: MW166213) based on Illumina pair-end sequencing. Fresh leaves from Wuyuan County, Jiangxi province, China (29°27′10.40″N, 117°44′16.65″E), were collected as sequencing materials. The voucher specimen (JXAU 2020026) was deposited in JXAU (the herbarium of the College of Forestry, Jiangxi Agricultural University, China. The Illumina paired-end (PE) library was prepared and sequenced in the Nanjing Novogene Biotechnology Co., Ltd. (Nanjing, China). The original reading was filtered by CLC Genomics Workbench version 9, and the clean reads were assembled by using GetOrganelle v1.5 (Jin et al. Citation2018). Subsequently, the plastome was annotated using GeSeq (Tillich et al. Citation2017) and Geneious 8.0.2 (http://www.geneious.com/), and sequences were aligned with MAFFT 7.409 (Katoh and Toh Citation2010).
The complete chloroplast genome of Styrax wuyuanensis is 157,969 bp in length, consisting of two inverted repeat regions with 25,954 bp (IRs), which were separated by a large single copy (LSC) with 87,575 bp, and small single copy (SSC) with 18,486 bp. The genome contained 132 functional genes, including 87 protein-coding, 37 tRNA, and eight rRNA. Most of the genes are single copy genes. However, seven protein-coding genes (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, and rps12), seven tRNA (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and four rRNA (rrn16, rrn23, rrn4.5, and rrn5) occur in duplicate. The overall GC content of the S. wuyuanensis chloroplast genome is 37.0%, with the corresponding values of LSC, SSC, and IR regions as 34.8%, 30.2%, and 43.0%, respectively.
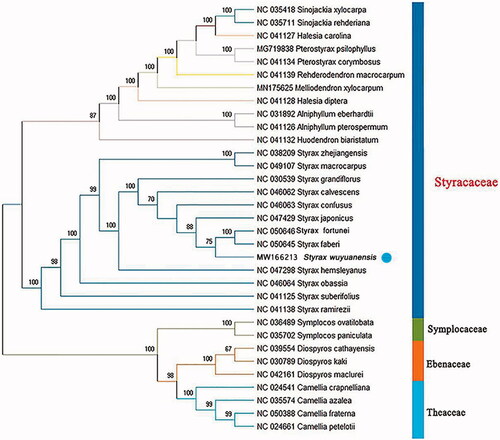
To evaluate the phylogenetic position of S. wuyuanensis, 23 Styracaceae chloroplast genomes together with three outgroup taxa in Symplocaceae, Ebenaceae, and Theaceae, separately, were downloaded from GenBank and analyzed. All sequences were aligned with MAFFT 7.409. The maximum-likelihood tree was constructed using RAxML 8.2.8 (Stamatakis Citation2014), using the GTR + Gamma substitution model and was based on 100 bootstrap replicates. Styrax wuyuanensis was positioned in a moderately supported clade (75%) with S. faberi and S. fortunei (). The molecular data confirm the morpho-anatomical evidence supporting the close relationship of S. wuyuanensis with the latter two species, both of which have the same fundamental morphological characters as S. wuyuanensis, i.e. the deciduous small shrub or small tree habit, the small (0.5–1.5 mm in diameter) and rounded fruit, and so on, in Styrax ser. Cyrta (Li and Fritsch Citation2018).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW166213. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA692531, SRR13447960, and SAMN17348586, respectively.
Additional information
Funding
References
- Hwang SM. 1980. A preliminary study on the family Styracaceae from China. Acta Phytotaxon Sin. 18(2):154–167.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479.
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26(15):1899–1900.
- Li G, Fritsch PW. 2018. A taxonomic revision of taxa in Styrax series Cyrta (Styracaceae) with valvate corollas. J Bot Res Inst Texas. 12(2):579–641.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45(W1):W6–W11.