Abstract
Viburnum sargentii Koehne is widely used for garden greening and also displays excellent medicinal value in China. However, the phylogenetic relationship between V. sargentii and other Adoxaceae members remains unknown. In this research, the complete chloroplast genome of V. sargentii was obtained by the high-throughput Illumina pair-end sequencing data. The chloroplast genome shows a typically quadripartite structure with 158,524 bp in size, including a large single-copy (LSC) region of 87,087 bp and a small single-copy (SSC) region of 18,489 bp, which were separated by two inverted repeat (IR) regions (26,474 bp each). A total of 128 genes were predicted, including 8 ribosomal RNAs (rRNAs), 37 transfer RNAs (tRNAs), and 83 protein-coding genes (PCGs). Maximum-likelihood phylogenetic analysis revealed that V. sargentii was clustered in the Viburnum genus and in a sister position to Viburnum japonicum, Viburnum erosum, Viburnum fordiae, and Viburnum betulifolium. This study provides useful information for future genetic study of V. sargentii.
Viburnum, was previously classified into the Caprifoliaceae, but the latest research shows that it belongs to the Adoxaceae (Clement et al. Citation2014). Viburnum sargentii Koehne belongs to the genus Viburnum, is widely distributed in the northeast and northwest region of China. V. sargentii is an excellent tree species for garden greening. It has a commonly known Chinese name as (‘Ji Shu Tiao’) and also used as Chinese herbal medicine. Many functional compounds (including flavonoids, terpenes, and phenylpropanoids) are isolated from V. sargentii (Bae et al. Citation2010; Xie et al. Citation2015); Modern pharmacological studies have shown that the 9′-O-methylvibsanol (isolated from V. sargentii) has a strong inhibitory effect on MCF-7 and A549 tumor cells (Bae et al. Citation2010). Although V. sargentii shows economic and medicinal value, the complete chloroplast genome of V. sargentii has not been reported. Here, in order to systematic understand the phylogenetic relationship of V. sargentii, we assembled and determined its complete chloroplast genome.
Fresh and Health leaves of V. sargentii were sampled from School of Traditional Chinese medicine, JILin Agriculture Science and Technology College, Jilin, Jilin Province, China (44°3′5.44″N, 126°6′34.44″E, 237 m above sea level). The leaf specimen (accession number: GL202002001) was deposited in the herbarium of School of Life Sciences, Guizhou Normal University (Jinghua Mu, [email protected]). The total genomic DNA (No. GL202002002) was extracted using DNAsecure Plant Kit (TIANGEN, Beijing, China) and stored at −80 °C in the laboratory (room number: 1403) of School of Life Science, Guizhou Normal University. A total concentration of 700 ng of DNA served as the input material for the DNA sample preparations. Sequencing libraries were generated using NEB Next® Ultra DNA Library Prep Kit for Illumina® (NEB, Ipswich, MA). The library preparations were sequenced on an Illumina platform and 150 bp paired-end reads were generated. After removal of adapter sequences, the clean filtered reads were assembled through the program GetOrganelle (Jin et al. Citation2019) with Viburnum erosum (GenBank accession number: MN641480) as the reference genome. The assembled chloroplast genome was annotated using the online software GeSeq) (Tillich et al. Citation2017). The accurate annotated complete chloroplast genome was submitted to GenBank under the accession number MW296955.
The length of circular complete chloroplast genome sequence of V. sargentii is 158,524 bp, consisting of a large single-copy (LSC, 87,087 bp) region and a small single-copy (SSC, 18,489 bp) region separated by two inverted repeat (IRA and IRB) regions of 26,474 bp each. In total, the chloroplast genome of V. sargentii comprised 128 distinct genes (predicted), including 83 protein coding (PCGs), 8 ribosomal RNA (rRNA), and 37 transfer RNA (tRNA) genes. Among the assembled genes, all rRNAs, 6 PCGs (rps2, rps7, rpl2, rpl23, ndhB, and ycf2), and 7 tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) occurred as double copies. Intron-exon analysis showed the majority (106 genes, 83%) of the genes with no introns, whereas 22 (17%) genes contained introns.
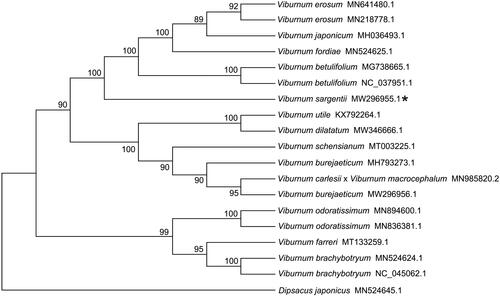
To further investigate the phylogenetic relationships between V. sargentii and other related species in Viburnum genus, 18 chloroplast genome sequences were downloaded from GenBank to construct a phylogenetic tree using maximum-likelihood (ML) method. These eighteen chloroplast genome sequences included seventeen species from Viburnum genus and one Caprifoliaceae member (Dipsacus japonicus) used as outgroup. The ML tree was performed using RAxML version 8.0.19 (Karlsruhe, Baden-Württemberg, Germany) (Model: GTRGAMMA) (Stamatakis Citation2014) with 1000 bootstrap replicates. The phylogenetic tree indicated that Viburnum japonicum and V. erosum first clustered together, and then Viburnum fordiae, Viburnum betulifolium, and V. sargentii clustered step by step (). Our complete chloroplast genome data of V. sargentii may contribute to a better understanding of the evolution and genetic studies of this plant.
Figure 1. Maximum likelihood tree based on the complete chloroplast genome sequences of 18 species in Viburnum genus shows the three distinct clusters. Dipsacus japonicus used as outgroup. GenBank accession numbers are described in the figure. Shown next to the nodes are bootstrap support values based on 1000 replicates.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession number MW296955. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682877, SRX9639379, and SAMN17013272, respectively.
Additional information
Funding
References
- Bae KE, Chong HS, Kim DS, Choi YW, Kim YS, Kim YK. 2010. Compounds from Viburnum sargentii Koehne and evaluation of their cytotoxic effects on human cancer cell lines. Molecules. 15(7):4599–4609.
- Clement WL, Arakaki M, Sweeney PW, Edwards EJ, Donoghue MJ. 2014. A chloroplast tree for Viburnum (Adoxaceae) and its implications for phylogenetic classification and character evolution. Am J Bot. 101(6):1029–1049.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv. 256479.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Xie Y, Wang J, Geng YM, Zhang Z, Qu YF, Wang GS. 2015. Pheonolic compounds from the fruits of Viburnum sargentii Koehne. Molecules. 20(8):14377–14385.
