Abstract
Photinia davidsoniae is a common ornamental arbor in the genus Photinia (family Rosaceae). Here, we sequenced and assembled the complete plastome of P. davidsoniae using the next-generation DNA sequencing technology. And we then compared it with nine Photinia species using a range of bioinformatics software tools. The ten plastomes had sizes ranging from 159,230 bp for P. beckii to 160,346 bp for P. davidsoniae. They all had a conservative quartile structure. It contained two single-copy regions: a large single-copy (LSC) region, a small single-copy (SSC) region, and a pair of inverted repeat (IR) regions. Each of the plastomes encoded 113 unique genes, including 79 protein-coding genes, four rRNA genes, and 30 tRNA genes. Furthermore, we detected six hypervariable regions (matK-rps16, rpoB-trnC, trnT-psbD, ndhC-trnV, psbE-petL, ndhF-rpl32-trnL), which could be used as potential molecular markers. We constructed two phylogenetic trees with plastomes or concatenated protein sequences of 25 species of 8 genera of Rosaceae. The tree constructed with complete plastomes has much stronger support. The results placed P. davidsoniae in the upper part of the phylogenetic tree. It shows that P. davidsoniae and P. lanuginosa are closely related. In summary, the plastomes of Photinia are conserved overall but carry significant minor variations, as expected. The results will be indispensable for distinguishing species, understanding the interspecific diversity, and elucidating the evolutionary processes of Photinia species.
1. Introduction
The genus Photinia belongs to Maleae (Rosaceae) and comprises approximately 60 species (Robertson et al. Citation1991; Lu and Spongberg Citation2003). They are widespread landscape tree species resistant to pruning and air pollution (Mattei et al. Citation2017; Mori et al. Citation2018), and many were cultivated for gardening (Zhao et al. Citation2020). Photinia davidsoniae Rehder & E.H.Wilson (referred to as P. davidsoniae in the following text) is an evergreen plant species, which grows in thickets at altitudes of 600–1000 m and mainly distributes in southern China and Southeast Asia (Lu and Spongberg Citation2003). Like many other species of Photinia, P. davidsoniae has luxuriant foliage around the trunk, purple and tender leaves in early spring, and little white flowers in early summer, and bear red fruits in autumn. (Sterling Citation1965; Aoki et al. Citation2006; Mattei et al. Citation2018). Photinia species exhibited similar morphological features and the species boundaries have been unclear. With the continuous discovery of new Photinia species (Guo et al. Citation2010; Li et al. Citation2015), a reliable classification of Photinia is in urgent need.
Chloroplast genomes (referred to as plastomes in the following text) have been widely used in plant taxonomy. Compared with morphological identification, plastome sequences can produce more accurate phylogenetic relationships. Recently, the confusion in the taxonomic of Photinia-related species has been primarily solved based on the complete plastome sequences (Shi et al. Citation2019; Liu, Liu, et al. Citation2019). Particularly, the phylogenetic analysis of the Photinia-related species support the idea of a new genus Phippsiomeles and the resurrection of a redefined Stranvaesia in Maleae (Liu, Hong, et al. Citation2019). Furthermore, Eriobotrya was found to belong to Rhaphiolepis based on plastomes and ribosome DNA (Liu, Liu, et al. Citation2019). However, their study focused only on the phylogenetic relationships among Photinia and its related genera. The comparative analysis of Photinia plastomes were not conducted extensively.
Here, we sequenced and assembled the complete plastomes of P. davidsoniae for the first time and then compared them with the plastomes of nine published Photinia species to explore the interspecific diversity of the plastomes of Photinia.
2. Materials and methods
2.1. Plant material, DNA extraction, and sequencing
We collected fresh leaves of P. davidsoniae from the Central China Medicinal Botanical Garden, EnShi, China (30°10′N, 109°44′E) and froze them at −80 °C. We used the plant genomic DNA kit (Tiangen Biotech, Beijing) to extract the total DNA following the manufacturer's protocol (Zhang, Li, et al. Citation2019). The DNA library with an insert size of 350 bp was constructed using the library preparation kit (New England Biolabs, USA) and sequenced using the Hiseq 2500 platform (Illumina, USA). We removed low-quality sequences, which are those with over 50% bases having quality values of Q < 19 or those with over 5% bases being ‘N.’ We obtained a total of 49,157,518 reads as clean data for further analysis.
2.2. Genome assembly and annotation
We used NOVOPlasty (v2.7.2) (Dierckxsens et al. Citation2017) to perform de novo genome assembly from the clean data. Bowtie2 (v2. 0.1) (Langmead et al. Citation2009) was used to ensure the assembly's correctness by mapping all clean reads to the assembled genome sequences. We used CPGAVAS2 (Shi et al. Citation2019) to annotate the genome. We used Apollo (Misra and Harris Citation2006) to edit the annotations with problems manually. The simple sequence repeats (SSRs) were identified using the CPGAVAS2 web server by calling MISA (Beier et al. Citation2017), including mono-, di-, tri-, tetra-, penta-, and hexanucleotides with the minimum numbers were 10, 5, 4, 3, 3, and 3, respectively. Additionally, tandem repeats were detected with the Tandem Repeats Finder program (v4.07b). REPuter (Kurtz et al. Citation2001) was used to calculate palindromic repeats, forward repeats, reverse repeats, and complementary repeats with the settings: Hamming Distance was three, and Minimal Repeat Size was 30 bp.
2.3. Analysis of comparative genomics and divergence hotspots
The plastomes of ten Photinia species, including other nine published plastomes: P. serratifolia (NC_045331.1), P. integrifolia (NC_045344.1), P. beckii (NC_045353.1), P. lanuginosa (NC_045354.1), P. prionophylla (NC_045355.1), P. lochengensis (NC_045352.1), P. glabra (MK920277.1), P. prunifolia (MK920279.1), and P. taishunensis (MK920278.1) were compared by using shuffle-LAGAN mode in mVISTA to identify interspecific variations (Thiel et al. Citation2003; Frazer et al. Citation2004). We conducted a sliding window analysis using DnaSP (v6.0) to calculate the nucleotide polymorphism (Pi) among the ten species. Lastly, IRscope (Amiryousefi et al. Citation2018) was used for visualizing the IR boundaries of the plastomes.
2.4. Phylogenetic analysis
The plastome sequences of 24 species in the family Rosaceae, including two outgroup species (Rosa rugosa and Sanguisorba officinalis), were downloaded from GenBank (Supplemental Table S1). The complete plastome sequences and 75 common protein sequences among the 25 species were aligned by using CLUSTALW2 (v2.0.1) (Thompson et al. Citation2002), respectively. These proteins include ACCD, ATPA, ATPB, ATPE, ATPF, ATPH, ATPI, CCSA, CEMA, CLPP, MATK, NDHA, NDHB, NDHC, NDHD, NDHE, NDHF, NDHG, NDHH, NDHI, NDHJ, NDHK, PETA, PETB, PETD, PETG, PETL, PETN, PSAA, PSAB, PSAI, PSAJ, PSBA, PSBB, PSBC, PSBD, PSBE, PSBF, PSBH, PSBJ, PSBK, PSBM, PSBN, PSBT, PSBZ, RBCL, RPL14, RPL16, RPL2, RPL20, RPL22, RPL23, RPL32, RPL33, RPL36,RPOA, RPOB, RPOC1, RPOC2, RPS11, RPS12, RPS14, RPS15, RPS16, RPS18, RPS19, RPS2, RPS3, RPS4, RPS7, RPS8, YCF1, YCF2, YCF3 and YCF4. These aligned sequences were used to construct the phylogenetic trees by using the Maximum Likelihood method (ML) implemented in RaxML (v8.2.4) (Stamatakis Citation2014). The parameters were ‘raxmlHPC-PTHREADS-SSE3 -f a -N 1000 -m PROTGAMMALGX/GTRGAMMA -x 551314260 -p 551314260’. The bootstrap analysis were performed with 1000 replicates.
3. Results
3.1. Basic features of the plastomes
The plastomes of Photinia are characterized by a typical circular DNA molecule with a total length ranged from 159,230 bp (P. beckii) to 160,346 bp (P. davidsoniae). The overall GC content ranged from 36.42% to 36.66%. These plastomes have a conservative quartile structure, comprising a large single-copy (87,434–88,302 bp) region, a small single-copy (19,217–19,361 bp) region, and a pair of inverted repeat (26,280–26,436 bp) regions (). The GC content of IR regions is higher than that of SSC regions and LSC regions in all ten Photinia species.
Table 1. Basic features of the plastomes of ten Photinia species.
3.2. Genome annotation
The genome structures of ten plastomes are highly conserved. Using the plastome of P. davidsoniae as an example, it contains 131 unique genes. Among them, 79 are protein-coding genes, four are rRNA genes, and 30 are tRNA genes ( and ). The total lengths of the protein-coding genes, rRNA genes, and tRNA genes are 77,832 bp, 9048 bp, and 2739 bp, accounting for 48.54%, 5.64%, and 1.71% of the complete plastome sequences, respectively. Introns play a significant role in selective gene splicing (Plangger et al. Citation2019). Among the 113 unique genes, two (ycf3 and clpP) contained two introns and 13 contained one intron, including eight protein-coding genes (rps16, atpF, rpoC1, petB, rpl22, rpl2, ndhB, ndhA) and five tRNA genes (trnK-UUU, trnS-CGA, trnL-UAA, trnE-UUC, trnA-UGC) (Table S2). We identified six protein-coding genes, four rRNAs genes, and seven tRNA genes duplicated in the IR regions. Three genes have been found to span the IR and single-copy regions, namely rps19, ndhF, and ycf1. Their structures are described in the section describing the contraction/expansion of the IR regions.
Figure 1. Graphic representation of features identified in P. davidsoniae plastome. The map was created with CPGAVAS2. Counted from the center, the first circle shows the forward and reverse repeats. They are connected with red and green arcs, respectively. The second and third circles indicate the locations of the tandem repeats and microsatellite sequences represented with short bars. The fourth circle shows the positions of plastome genes. The genes are colored base on their functional categories.
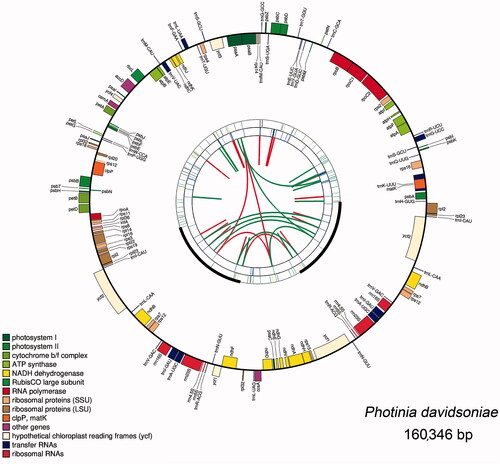
Table 2. Gene contents of the plastomes of Photinia species.
3.3. Repeats analysis
In this study, the numbers of SSRs ranged from 95 (P. prionophylla) to 105 (P. taishunensis). And we detected 1001 SSR loci in the ten plastome sequences (). It is worth noting that most of the repeating units in Photinia plastomes were A/T repeats, resulting in high A/T content in these plastomes. Besides, trinucleotide repeats are rare, and we only observed one in P. prunifolia.
Figure 2. Comparison of the Repeats in the plastomes of Photinia species. (A) Types and numbers of SSRs detected in the plastomes of ten Photinia species; (B) Types and numbers of tandem repeats and dispersed repeats detected in the plastomes of ten Photinia species.
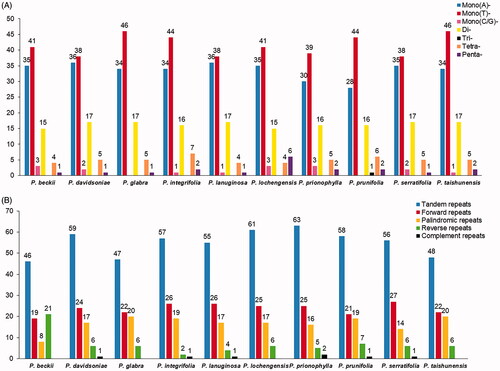
Moreover, we detected 550 tandem repeats in the ten cp genomes using the similarity cutoff of 90% (). The numbers of tandem repeats ranged from 46 (P. beckii) to 63 (P. prionophylla). In contrast, the length of tandem repeats is mostly <= 20 bp (data are not shown). Besides, 237 forward repeats, 167 palindromic repeats, 69 reverse repeats, and seven complementary repeats were detected. Moreover, we found that P. beckii had the largest number of reverse repeats and the least number of palindromic repeats, different from the other nine species ().
3.4. Contraction and expansion of the IR regions
With the evolution of plastomes, the IR regions have expanded and contracted, and some genes have the opportunity to access the IR regions or single-copy regions (Wang et al. Citation2018). The IR regions can undergo contraction and expansion, which are considered the main reason for the different lengths of plastomes in angiosperms. We compared the IR and SC boundaries of ten Photinia species and five related genera species (). Three genes, rps19, ndhF, and ycf1, were found to span the borders. Most rps19 sequence is in the LSC region, and only a small fragment is in the IRb region. The length of the small fragment varies significantly. For example, it is 2 bp for P. taishunensis and P. glabra, 28 bp for P. beckii. In contrast, it is over 100 bp in other species.
Figure 3. Comparison of LSC, SSC, and IR border regions of species from Photinia and related genera. The genes around the borders are shown above or below the mainline. The JLB, JSB, JSA, and JLA represent junction sites of LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC, respectively. ‘*’: pseudogene.
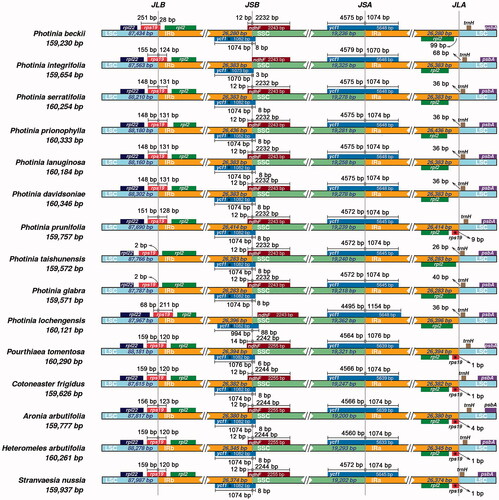
For gene ndhF, they are located in the IRb/SSC border and overlapping with the first copy of the ycf1 gene. The large fragment of ndhF is in the SSC region; the small segment of ndhF is in the IRb region. The exception is that the ndhF genes for P. integrifolia and P. lochengensis are completely included in the SSC regions. For gene ycf1, there have two copies, which span the junctions of IRb/SSC and SSC/IRa. The length of the fragments in the IRa and IRb regions are similar, about 1000 bp.
3.5. Genome divergence and hypervariable regions
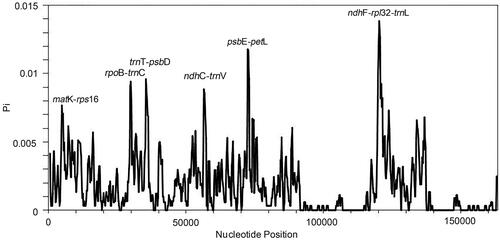
To evaluate the genomic divergence, we analyzed ten plastomes using mVISTA. The LSC and SSC regions showed a high divergence level than the IR regions, particularly for the non-coding regions. Three non-coding regions showed significant divergence: trnR-atpA, trnT-psbD, and ndhC-trnV (). Overall, most regions of the analyzed sequences showed a high degree of similarity. To quantify the levels of DNA polymorphism, the ten plastomes were analyzed by using DnaSP (v6.0) and we detected six hypervariable regions: matK-rps16 (0.00767), rpoB-trnC (0.00941), trnT-psbD (0.00959), ndhC-trnV (0.00885), psbE-petL (0.01178), and ndhF-rpl32-trnL (0.01385). The Pi values are listed in the parentheses. All of them are intergenic regions (). These hypervariable regions could be used as potential molecular markers.
Figure 4. Comparison of the cp genomes in Photinia species by using mVISTA. The gray arrows on the top of the alignment represent genes. The pink regions are ‘Conserved Non-Coding Sequences’ (CNS), the dark blue regions are exons, and the light-blue regions are tRNA or rRNA. The percentages (50% and 100%) are the similarity among these sequences. Gray arrows on the top of the aligned sequences represent genes and their orientation. The colored lines under each window represent different regions of the plastomes: the blue line refers to the LSC region, the red line refers to the IR regions, and the green line refers to the SSC region.
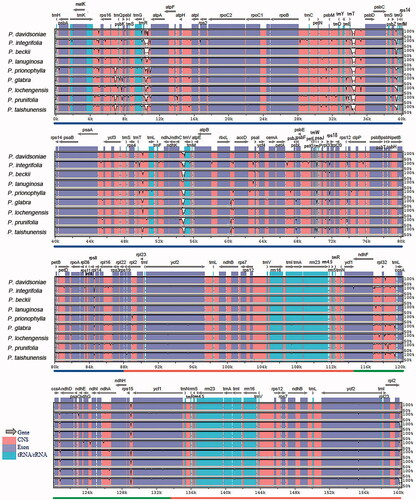
Figure 5. Hypervariable regions in Photinia species. We used a sliding window to analyze the sequence polymorphism among the plastomes of ten Photinia species. The sliding window has a length of 600 bp and a step size of 200 bp. The X-axis represents the position of nucleotide; Y-axis represents nucleotide polymorphism of each window.
3.6. Phylogenetic analysis using plastomes data
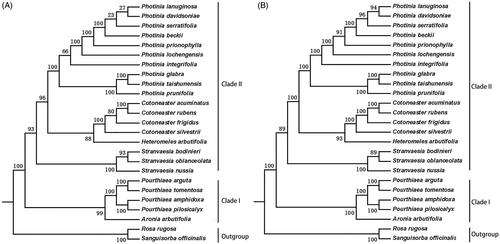
Here, we use two data sets to determine the phylogenetic relationship: the common protein-coding sequences () and the complete plastome sequences (). In our phylogenetic trees, it is evident that there are two main clades and then further divided into different subclades. Clade I included Pourthiaea and Aronia, and clade II contained four genera: the Photinia, Heteromeles, Cotoneaster, and Stranvaesia. Our data showed that P. davidsoniae is most closely related to P. lanuginosa. Both were most closely related to P. serratifolia. The two phylogenetic trees have similar topologies. However, the branches from the tree constructed with the complete chloroplast genome sequences have higher bootstrap support values. It is possible that the protein sequences are highly conserved and thus do not have sufficient informative sites to determine the relationships among these species.
Figure 6. Phylogenetic relationships of species from Photinia and related genera inferred using the Maximum likelihood (ML) method. A. The phylogenetic tree was constructed was constructed using the complete nucleotide sequences of the using the 75 common protein sequences among the 25 cp genomes. B. The phylogenetic tree 25 cp genomes. Two taxa, namely, R. rugosa and S. officinalis, were used as outgroups. Bootstrap values were calculated from 1000 replicates.
4. Discussion
In this study, we sequenced the plastome of P. davidsoniae to understand its phylogenetic relationship with other congeneric species and also carried out a detailed comparative analysis of ten plastomes from Photinia.
The plastomes were found to be highly conserved from different aspects. For example, these plastomes have identical numbers of protein-coding genes, rRNA genes, and tRNA genes. There is no rearrangement among the plastomes, consistent with those described for most other genera in angiosperms (Raman et al. Citation2017). Furthermore, our phylogenetic results are consistent with the earlier investigation (Shi et al. Citation2019; Liu, Liu, et al. Citation2019). And the results show that the whole plastome sequences are more reliable in phylogenetic and evolutionary studies as a super barcode (Zhang, Zhang, et al. Citation2019).
Nevertheless, we have identified minor variations among these plastomes. Firstly, there are differences in the number of repeats detected in different species, including SSRs, tandem repeats, and dispersed repeats. SSRs exhibited high polymorphism in Photinia species, which have provided a large amount of information for molecular markers (Wang et al. Citation2017). Previous research reported that these short dispersed repeats ranged from 30 to 40 bp are essential for promoting plastomes rearrangements. Whether these repeats have caused the rearrangement of the cp genomes of Photinia species is an interesting question.
Secondly, changes were observed in the IR boundary region of these plastomes. Although these changes are subtle, two genes, rps19 and ndhF, exhibited significantly dynamic changes at LSC/IRb and IRb/SSC. For gene rps19, it didn’t span the LSC/IRb border in two species (P. taishunensis and P. glabra). Similarly, ndhF genes did not span the IRb/SSC boundary in some species (P. integrifolia and P. lochengensis). However, both genes span the boundaries in most cases. It is not clear whether this dynamic boundary change has any effect on these genes’ transcription.
Lastly, there were high nucleotide polymorphisms in the non-coding region of the plastomes. Some intergenic regions are potentially hypervariable regions for the development of molecular markers. We recommend six hypervariable regions, matK-rps16 (0.00767), rpoB-trnC (0.00941), trnT-psbD (0.00959), ndhC-trnV (0.00885), psbE-petL (0.01178), and ndhF-rpl32-trnL (0.01385), as potential molecular markers. These markers have important applications for rapid interspecific identification of Photinia taxa.
We learned several lessons from these studies. Firstly, for closely related species, the non-coding regions might provide useful information to understand the current evolutionary processes. Secondly, one needs to incorporate additional information from the nuclear genomes and mitochondrial genomes for overall phylogenetic and evolutionary analysis. Unfortunately, we could not retrieve the raw sequence data for these analyses. For the future, more genome sequencing is needed to further explore these issues.
In summary, the results reported here could provide valuable information for genetic diversity, phylogenetic evolution, and taxonomy studies of the genus Photinia.
Authors’ contributions
CL and JY conceived the study. LQ.W. collected the samples. HM.C .extracted DNA for sequencing by using Illumina and PacBio technologies. MJ assembled the genome, and JLL annotated the genome. Next, MJ and JTL carried out the repeat analysis and phylogenetic analysis. Finally, JLL wrote the paper. All authors have read and agreed on the contents of the manuscript.
Acknowledgments
The authors are grateful to Prof. Jingwen You from Central China Medicinal Botanical Garden for providing sample resources.
Disclosure statement
The authors declare no conflict of interest. The funders have not participated in the study design, data collection, analysis, interpretation, and manuscript writing. They are not involved in the decision to publish the results.
Data availability statement
The sample has been deposited in the herbarium of the Institute of Medicinal Plant Development in Beijing, China, with the accession number: implad201808128. The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT230547. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA688534, SAMN17180600, and SRR13325884, respectively.
Additional information
Funding
References
- Amiryousefi A, Hyvonen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34(17):3030–3031.
- Aoki K, Matsumura T, Hattori T, Murakami N. 2006. Chloroplast DNA phylogeography of Photinia glabra (Rosaceae) in Japan. Am J Bot. 93(12):1852–1858.
- Beier S, Thiel T, Munch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32(Web Server issue):W273–279.
- Guo W, Yu Y, Fan Q, Ma J-S, Liao W-B. 2010. “Photinia sorbifolia” (Rosaceae), a new species from Hunan Province, China. Annales Botanici Fennici. 47(5):394–396.
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29(22):4633–4642.
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.
- Li Q, Wu Y, Lu D-D, Xu Y-F, Lin Y-R. 2015. A new species of Terriera (Rhytismatales, Ascomycota) on Photinia villosa. Mycotaxon. 130(1):27–31.
- Liu BB, Hong DY, Zhou SL, Xu C, Dong WP, Johnson G, Wen J. 2019. Phylogenomic analyses of the Photinia complex support the recognition of a new genus Phippsiomeles and the resurrection of a redefined Stranvaesia in Maleae (Rosaceae). Jnl of Sytematics Evolution. 57(6):678–694.
- Liu BB, Liu GN, Hong DY, Wen J. 2019. Eriobotrya belongs to Rhaphiolepis (Maleae, Rosaceae): evidence From Chloroplast Genome and Nuclear Ribosomal DNA Data. Front Plant Sci. 10:1731.
- Lu LT, Spongberg SA. 2003. Photinia lindley. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 9. Beijing: Science Press; St Louis: Missouri Botanical Garden Press; p. 46–434.
- Mattei P, D'Acqui LP, Nicese FP, Lazzerini G, Masciandaro G, Macci C, Doni S, Sarteschi F, Giagnoni L, Renella G. 2017. Use of phytoremediated sediments dredged in maritime port as plant nursery growing media. J Environ Manage. 186(Pt 2):225–232.
- Mattei P, Gnesini A, Gonnelli C, Marraccini C, Masciandaro G, Macci C, Doni S, Iannelli R, Lucchetti S, Nicese FP, et al. 2018. Phytoremediated marine sediments as suitable peat-free growing media for production of red robin photinia (Photinia x fraseri). Chemosphere. 201:595–602.
- Misra S, Harris N. 2006. Using Apollo to browse and edit genome annotations. Curr Protoc Bioinformatics. Chapter 9:Unit 9.5.
- Mori J, Fini A, Galimberti M, Ginepro M, Burchi G, Massa D, Ferrini F. 2018. Air pollution deposition on a roadside vegetation barrier in a Mediterranean environment: combined effect of evergreen shrub species and planting density. Sci Total Environ. 643:725–737.
- Plangger R, Juen MA, Hoernes TP, Nußbaumer F, Kremser J, Strebitzer E, Klingler D, Erharter K, Tollinger M, Erlacher MD, et al. 2019. Branch site bulge conformations in domain 6 determine functional sugar puckers in group II intron splicing. Nucleic Acids Res. 47(21):11430–11440.
- Raman G, Park V, Kwak M, Lee B, Park S. 2017. Characterization of the complete chloroplast genome of Arabis stellari and comparisons with related species. PLoS One. 12(8):e0183197.
- Robertson KR, Phipps JB, Rohrer JR, Smith PG. 1991. A synopsis of genera in Maloideae (Rosaceae). Syst Bot. 16(2):376–394.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sterling C. 1965. Comparative morphology of the carpel in the Rosaceae VI. Pomoideae: Eriobotrya, Heteromeles, Photinia, Pourthiaea, Raphiolepis, Stranvaesia. Am J Bot. 52(9):938–946.
- Thiel T, Michalek W, Varshney RK, Graner A. 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet. 106(3):411–422.
- Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. Chapter 2:Unit 2.3.
- Wang W, Chen S, Zhang X. 2018. Whole-genome comparison reveals divergent IR borders and mutation hotspots in chloroplast genomes of herbaceous bamboos (Bambusoideae: Olyreae). Molecules. 23(7):1537.
- Wang Z, Zeng Y, Zhang Z, Sheng S, Tian J, Wu R, Pang X. 2017. Phylogeography study of the Siberian apricot (Prunus sibirica L.) in Northern China assessed by chloroplast microsatellite and DNA makers. Front Plant Sci. 8:1989.
- Zhang M, Li L, Li B, Tian N, Yang M, Zhang H, You C, Zhang J. 2019. Adsorption of DNA by using polydopamine modified magnetic nanoparticles based on solid-phase extraction. Anal Biochem. 579:9–17.
- Zhang Z, Zhang Y, Song M, Guan Y, Ma X. 2019. Species identification of dracaena using the complete chloroplast genome as a super-barcode. Front Pharmacol. 10:1441.
- Zhao D, Liu G, Wang X, Daraz U, Sun Q. 2020. Abundance of human pathogen genes in the phyllosphere of four landscape plants. J Environ Manage. 255:109933.
