Abstract
Exostoma gaoligongense is an endemic glyptosternine catfish distributed in the Nujiang River drainage, Yunnan Province, China, with few published genetic information. In this study, we sequenced and characterized the complete mitochondrial genome of E. gaoligongense, which was circular, 16529 bp in length, containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, one replication origin and one control region. Phylogenetic analysis revealed that E. gaoligongense had the closest relationship with its congener E. labiatum. They clustered with the clade containing most genera of glyptosternine catfishes and then cluster with the more primitive genera Glaridoglanis and Glyptosternon.
The genus Exostoma Blyth 1860, belonging to the glyptosternine catfishes within Sisoridae, is characterized by having a strongly depressed body profiles and horizontally enlarged paired fins modified for adhesion, continuous post-labial groove, homodont dentition consisting of distally flattened oar-shaped teeth and two narrowly-separated patches on the upper jaw and two well-separated patches on the lower jaw (Thomson and Page Citation2006; Darshan et al. Citation2019). Species of genus Exostoma primarily inhabit the bottom of the rocks in the swift-flowing mountain streams or rivers from the Yarlung Tsangpo-Brahmaputra River drainage eastwards to the Lancang-Mekong River drainage (Gong et al. Citation2018). To date, 18 valid species within this genus have been described (Luo and Chen Citation2020; Arunkumar Citation2020). E. gaoligongense, recently described by Chen et al. (Citation2017) based on morphological diagnosis, was endemic to four tributaries (Manggang River, Tangxi River, Xinya River and Nanzha River) of Nujiang (upper Salween) River in Yunnan, China.
In this study, a female specimen of E. gaoligongense (52 mm standard length) was collected during June, 2020 from Kungong River, a tributary of Nujiang River in Lujiang Town, Baoshan City, Yunnan (new record of distribution, 25°5′40.9″N, 98°49′18.6″E, 797 m), which were deposited at the Kunming Natural History Museum of Zoology, Chinese Academy of Sciences (voucher number: KIZ 2020000021). The genome DNA was extracted from a bit of fin tissue using the Animal Tissue DNA Extraction Kit (TSP201, Tsingke, China). Paired-end sequencing for the mitogenome of E. gaoligongense was performed on the Illumina Hiseq platform (Illumina Inc., San Diego, CA). The obtained raw sequence reads were assembled into contigs using SPAdes 3.9 (Bankevich et al. Citation2012) and the annotation was conducted in MITOS (Bernt et al. Citation2013).
The complete mitogenome sequence of E. gaoligongense is circular and 16529 bp in length, including 13 protein-coding genes, 22 tRNA genes, two rRNA genes, one replication origin (OL) and one control region (D-loop). The total nucleotide composition of E. gaoligongense mitogenome is 32.4% A, 28.7% C, 24.3% T, and 14.6% G. The high A-T content pattern is consistent with the typical feature of the vertebrate mitochondrial genome (Mayfield and McKenna Citation1978). Most of the protein-coding genes started with ATG as the initiation codon except for CO1 gene starting with GTG. ND4, COX2, COX3, and Cytb genes ended with T- whereas the remaining 9 protein-coding genes ended with ATR (R represents A or G). Structure of protein-coding genes in E. gaoligongense was identical to that in its congener E. labiatum (NC021601).
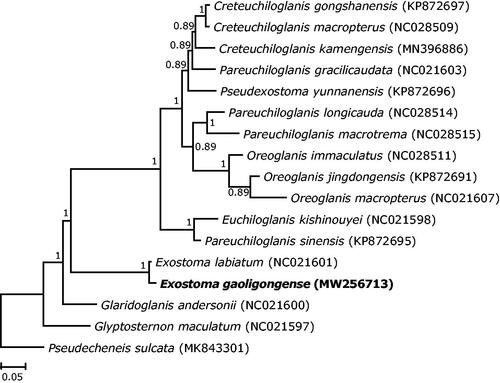
The 13 mitochondrial protein-coding genes were combined from E. gaoligongense and 16 additional species including 15 glyptosternine species and one outgroup (Pseudecheneis sulcatus) for phylogenetic analysis. Sequence alignment was performed using MEGA X 10.0 (Kumar et al. Citation2018). Nucleotide substitution model (GTR + I + G) was chosen based on Akaike information criterion (AIC) using jModelTest 2.1 (Darriba et al. Citation2012). Phylogenetic analysis was performed based on Bayesian inference in MrBayes 3.1 (Ronquist and Huelsenbeck Citation2003).
The topology of the phylogenetic tree showed that E. gaoligongense clustered with its congener E. labiatum (NC021601, location: Nujiang River) with small genetic divergence of 1.6% Kimura 2-parameter distance. In addition, Exostoma species clustered with the clade containing the most genera of glyptosternine catfishes and then clustered with the more primitive groups Glaridoglanis and Glyptosternon (). The mitogenome announcement of E. gaoligongense could help understand the phylogenetic position of this genus, as well as offer valuable genetic information for further study on the genetic diversity and biological conservation of this species.
Discloser statement
All authors declare no potential conflict of interest.
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/1945652357.
Additional information
Funding
References
- Arunkumar L. 2020. Exostoma laticaudata, a new glyptosternine catfish (Teleostei: Sisoridae) from Manipur, northeastern India. Species. 21(68):293–305.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Externbrink F, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen XY, Poly WJ, Catania D, Jiang WS. 2017. A new species of sisorid catfish of the genus Exostoma from the Salween drainage, Yunnan, China. Zool Res. 38(5):291–299.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Darshan A, Vishwanath W, Abujam S, Das DN. 2019. Exostoma kottelati, a new species of catfish (Teleostei: Sisoridae) from Arunachal Pradesh, India. Zootaxa. 4585(2):369.
- Gong Z, Lin P, Liu F, Liu H. 2018. Exostoma tibetana, a new glyptosternine catfish from the lower Yarlung Tsangpo River drainage in southeastern Tibet, China (Siluriformes: Sisoridae). Zootaxa. 4527(3):392–402.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Luo ZJ, Chen XY. 2020. Exostoma dulongensis, a new glyptosternine catfish from the Irrawaddy basin, Yunnan, China (Siluriformes: Sisoridae). Zootaxa. 4802(1):99–110.
- Mayfield JE, McKenna JF. 1978. A-T rich sequences in vertebrate DNA. A possible explanation of q-banding in metaphase chromosomes. Chromosoma. 67(2):157–163.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1755.
- Thomson AW, Page LM. 2006. Genera of the Asian catfish families Sisoridae and Erethistidae (Teleostei: Siluriformes). Zootaxa. 1345(1):1–96.