Abstract
The mitochondrial genome (mitogenome) of Nothybus sumatranus is described in the present paper, representing the first mitogenome reported from the family Nothybidae. The nearly complete mitochondrial genome is 16,128 bp in length, containing 13 protein-coding genes, 22 tRNA genes, and two rRNA genes. Genome organization, nucleotide composition and codon usage of the mitogenome are noted, and secondary structures of all tRNAs are predicted. The sister relationship between Diopsoidea and Nerioidea was supported by the phylogenetic tree based on Bayesian inference (BI) method.
The family Nothybidae belongs to the superfamily Diopsoidea of Acalyptratae (Diptera). It is a small group currently contains only twelve species in the single genus Nothybus Rondani, 1875 (Lonsdale Citation2020). Nothybids are slender, elongate, long-legged flies ranging from 5.5 to 15.0 mm in size, usually yellowish orange to brown with abdomen partially dark brown to blackish brown (Zhou et al. Citation2021). Members of Nothybidae are mainly distributed in the Oriental Region (Lonsdale and Marshall Citation2016). Enderlein (Citation1922) described an Indonesian species named Nothybus sumatranus Enderlein, Citation1922, which has also been recorded from China, Malaysia, Thailand and Vietnam (Lonsdale and Marshall Citation2016; Zhou et al. Citation2021). This species can be distinguished from its congeners by the combination of the following characters: frons with two velvety black anterior patches; mesonotum with irregularly eight rows of presutural acrostichal setulae in mid-longitudinal stripe; wing with three clear iridescent spots with second spot in cell r4 + 5 displaced apically, crossvein r-m with narrow medial brown spot; subapical spot on CuA1 absent; fore basitarsomere white with base dark brown; abdominal tergite I and anterolateral corner of tergite II widely yellowish-orange, tergite III laterally with orange regions (Zhou et al. Citation2021). In this study, the mitogenome of N. sumatranus is sequenced and described as the first mitogenome reported from the family Nothybidae.
Voucher specimen of N. sumatranus (No. Zc-DN001) was collected from Cat Tien National Park, Dong Nai, Vietnam (N11.45799°, E107.31972°) and deposited in the Entomological Museum of China Agricultural University, Beijing, China (CAU, Ding Yang, [email protected]). The genomic DNA was extracted from thoracic muscle tissues using DNeasy DNA Extraction kit (TIANGEN) and stored under −20 °C. The mitogenome was sequenced using the Illumina Hiseq 2500 platform with 150 bp paired-end reads. Adapters and low quality and short reads were removed using Trimmomatic (Bolger et al. Citation2014) and Prinseq (Schmieder and Edwards Citation2011), respectively. A total of 4 Gb clean data were obtained and used in the de novo assembly using IDBA-UD (Peng et al. Citation2012), with minimum and maximum k values of 80 and 240 bp. The assembly accuracy was checked by Geneious 10.1.3 (http://www.geneious.com/). This produced the partial mitogenome of N. sumatranus with an average sequencing depth of 54.7×. The sequence was deposited in GenBank under the accession number MW387954.
The nearly complete mitogenome of N. sumatranus (16,128 bp) is obtained, including 37 genes (13 protein-coding genes, 22 tRNA genes, and two rRNA genes) and a partial control region. Gene order is the same with other sequenced Acalyptratae fly mitogenomes reported before, without rearrangement occurring in this mitogenome (Clary and Wolstenholme Citation1985; Li et al. Citation2019, Citation2020). The nucleotide composition of this mitogenome is 39.2% of A, 38.9% of T, 8.7% of G, 13.2% of C, and A + T content is 78.1% of the entirety. All of 13 protein-coding genes initiate with ATN as the start codon (six with ATG, three with ATT, two with ATC, and one with ATA), except COI used TCG as its start codon. The stop codon TAA and TAG are assigned to seven and two protein-coding genes, respectively, whereas a single T residue is used by COII, NAD1, NAD4 and NAD5 as an incomplete stop codon. The length of tRNA genes ranges from 62 to 72 bp. All tRNA genes can be folded into the typical clover-leaf secondary structure. The lrRNA is 1,321 bp in length with A + T content of 82.8%, and the srRNA is 787 bp long with A + T content of 81.7%.
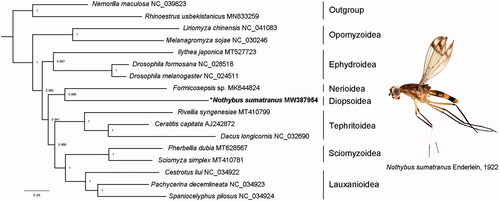
Phylogenetic tree including 15 Acalyptratae fly species and two outgroups was reconstructed by Bayesian inference (BI) method (). The sister relationship between Diopsoidea (represented by N. sumatranus) and Nerioidea (represented by Formicosepsis sp.) was supported with relatively high Bayesian posterior probabilities (BPP = 0.986), which was consistent with some previously hypothesis (McAlpine Citation1989; Marshall Citation2012). The phylogenetic topology also recovered the sister relationship between Opomyzoidea and the remaining Acalyptratae superfamilies (BPP = 1), while the latter formed the relationship as Ephydroidea + ((Diopsoidea + Nerioidea) + (Tephritoidea + (Lauxanioidea + Sciomyzoidea))). The mitogenomic data of N. sumatranus could provide basic genetic information for future phylogenetic and evolutionary studies of the family Nothybidae and the superfamily Diopsoidea.
Figure 1. Phylogenetic relationship of 15 acalyptrate fly species inferred from BI analysis based on 13 protein-coding genes. Phylogenetic tree was generated by MrBayes (Ling et al. Citation2016). Asterisk indicates the newly sequenced data in this study. GenBank accession numbers of all sequence used in the phylogenetic tree have been included in the figure.
Disclosure statement
All authors have read and approved the final manuscript. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The mitogenome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, under the accession number MW387954. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA716164, SRR13908658, and SAMN18396143, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22(3):252–271.
- Enderlein G. 1922. Klassifikation der Micropeziden. Arch Naturgesch (A). 88:140–229.
- Ling C, Luo A, Gao J. 2016. MrBayes 3.2.6 on Tianhe-1A: a high performance and distributed implementation of phylogenetic analysis. IEEE 22nd International Conference on Parallel and Distributed Systems (ICPADS). p. 1181–1186.
- Li X, Wang L, Li Z, Hou P, Yang D. 2019. The mitochondrial genome of Formicosepsis sp. (Diptera: Cypselosomatidae). Mitochondrial DNA B Resour. 4(2):2140–2141.
- Li X, Zhou J, Wang L, Hou P, Yang D. 2020. The mitochondrial genome of Prosthiochaeta sp. (Diptera: Platystomatidae). Mitochondrial DNA B Resour. 5(3):2557–2558.
- Lonsdale O, Marshall SA. 2016. Revision of the family Nothybidae (Diptera: Schizophora). Zootaxa. 4098(1):1–42.
- Lonsdale O. 2020. Family groups of Diopsoidea and Nerioidea (Diptera: Schizophora)—Definition, history and relationships. Zootaxa. 4735(1):1–177.
- Marshall SA. 2012. Flies: the natural history and diversity of Diptera. Richmond Hill (Ontario): Firefly Books Ltd; p. 1–616.
- McAlpine JF. 1989. Phylogeny and classification of the Muscomorpha. In: McAlpine JF. Manual of the Nearctic Diptera. Vol. 3. Ottawa: Biosystematics Research Institute; p. 1397–1518.
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428.
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 27(6):863–864.
- Zhou J, Marshall SA, Yang D. 2021. Synopsis of Nothybidae (Diptera: Brachycera) from China, with redescription of Nothybus absens Lonsdale & Marshall. Zootaxa. 4926(2):201–223.
