Abstract
The complete chloroplast genome of Mucuna sempervirens reported herein was a circular DNA molecule of 154,542 bp in length. The genome had a typical quadripartite structure, consisting of a pair of inverted repeats (IRa and IRb: 24,836 bp) separated by a large single-copy region (LSC: 67,996 bp) and a small single-copy region (SSC: 18,363 bp). The overall GC content of the genome was 35.1%. The cp genome encoded a set of 128 genes, containing 82 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis indicated that M. sempervirens was sister to M. macrocarpa. These findings may provide useful information to the phylogeny of the genus Mucuna.
Mucuna sempervirens Hemsl is a lianas evergreen species belong to the genus Mucuna (Fabaceae: Papilionoideae). They grow in subtropical forests, bushes, valleys, and rivers, which are widely distributed in northwest China (Sichuan, Guizhou, Yunnan, Shaanxi, Hubei, Zhejiang, Jiangxi, Hunan, Fujian, Guangdong, and Guangxi), and Japan. It has a high value of ornamental and medicinal application (Du and Li Citation2012).
Fresh leaves of M. sempervirens were sampled from Lianfeng peak, Jiuhuashan Mountain, Anhui Province, China (30.51 N, 117.85E, altitude 236 m). The voucher specimen was preserved and deposited in the Museum of Huangshan University (Voucher number: YMT2021; Contact person: ZB Wan and email: [email protected]). Total DNA extraction and whole-genome sequencing were conducted by Nanjing Genepioneer Biotechnologies Inc. (Nanjing, China) with the Illumina Hiseq X Ten platform. Raw data was filtered using the fatap 0.20.0 (Chen et al. Citation2018). A total of 20,341,388 clean reads were accomplished by CLC v9.11 (Nicolas et al. Citation2017). The alignment of contigs was under the BLAST algorithm (Johnson et al. Citation2008) with M. macrocarpa (NC044116) plastid genome as reference. The genome was annotated using the CpGAVAS pipeline (Liu et al. Citation2012) and submitted to GenBank (Accession number: MW556306). MsatCommander v0.8.2.0 (Faircloth, Citation2008) was utilized to identify simple sequence repeats (SSRs).
The complete cp genome of M. sempervirens was 154,542 bp in length and exhibited a typical quadripartite structure, which was composed of a pair of inverted repeats (IRa and IRb: 24,836 bp) separated by a large single-copy region (LSC: 67,996 bp) and a small single-copy region (SSC: 18,363 bp). The overall GC content of the genome was 35.1%, while the corresponding values of LSC, SSC, and IR regions were 32.9%, 30.5%, and 42.3%, respectively. A total of 128 genes were annotated in the cp genome, including 82 protein-coding genes, 37 tRNA genes, and eight rRNA genes.
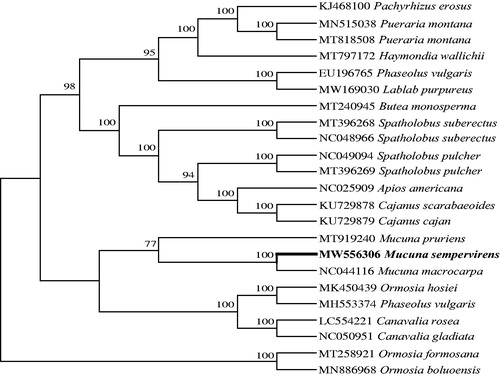
To investigate phylogenetic position of M. sempervirens, the sequence alignment was performed on the 24 cp genome sequences using MAFFT 7.307 (Katoh et al. Citation2019), and then jModelTest v2.1.7 (Darriba et al. Citation2012) was utilized to decide the optimal model, including 22 Phaseoleae species and 2 Ormosieae species as outgroup. Maximum likelihood (ML) method was used to reconstruct phylogenetic tree by online RAxML BlackBox software in http://www.phylo.org (Stamatakis et al. Citation2008). The phylogenetic analysis result shows that M. sempervirens is sister to M. macrocarpa and resides within the clade representing the genus Mucuna () with highly bootstrap support values. It indicated that our new determined complete cp genome sequence could meet the demands and explain some evolution issues.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome sequence and annotation of M. sempervirens that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MW556306.
Additional information
Funding
References
- Chen SF, Zhou YP, Chen YR, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Du Q, Li B. 2012. Identification of antioxidant compounds of Mucuna sempervirens by high-speed counter-current chromatographic separation–dpph radical scavenging detection and their oestrogenic activity. Food Chem. 131(4):1181–1186.
- Faircloth BC. 2008. Msatcommander: detection of microsatellite repeat arrays and automated, locus-specific primer design . Mol Ecol Resour. 8(1):92–94.
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36(Web Server issue):W5–W9.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Nicolas D, Patrick M, Guillaume S. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57(5):758–771.