Abstract
In this study, the whole chloroplast genome of Ulmus bergmanniana was sequenced, assembled, annotated and characterized for the first time. The results showed that the total length of U. bergmanniana chloroplast genome was 160,060 bp and the GC content was 35.52%, which had the typical circular double-stranded tetrad structure of angiosperm chloroplast genome. It includes a large single-copy region (LSC), a small single-copy region (SSC), and a pair of inverted repeat regions (IRa/IRb), with lengths of 88,161 bp, 19,029 bp, and 26,435 bp, respectively. A total of 131 genes were annotated, including 86 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Based on the chloroplast genome sequences of 14 plants in U. bergmanniana and NCBI database, the phylogenetic tree was constructed, and the phylogenetic position of U. bergmanniana in Ulmaceae was determined. The results will provide a theoretical basis for molecular identification and resource development and utilization of U. bergmanniana.
There are nearly 230 species of Ulmaceae plants in 16 genera. They are widely distributed in temperate and tropical regions of the world. They mainly produce the northern temperate zone. Taxonomists have always been in disagreement on the location of its system evolution, the location of the system and the determination of species (Chernik Citation1981; Zavada and Crepet Citation1981; Zavada Citation1983; Tobe and Takaso Citation1990). Because the chloroplast genome is more conservative than the nuclear genome and the mitochondrial genome in terms of genome size, genome structure and gene content (Palmer Citation1985), the genes obtained from the chloroplast genome have been used to distinguish species and phylogenetic analysis (Gu et al. Citation2019). Recently, studies on chloroplast genomes and phylogeny of Ulmus have been carried out in Ulmaceae, such as U. elongata and U. lanceaefolia (Ebrahimi et al. Citation2020; Shuxiang et al. Citation2020; Yinran et al. Citation2020), but there is no related report on U. bergmanniana.
U. bergmanniana belongs to deciduous tree. Distributed in Gansu Province, Shanxi Province and so on in China. It is often born in broad-leaved forests on mountain slopes and streams at an altitude of 1500-2600 meters. The test materials were planted in Shijiazhuang, China (114°28′12″E, 38°08′23″N). The experimental material was stored in herbarium of Hebei Academic of Forestry and Grassland, File number is HAFG22U354. Total DNA of fresh young leaves was extracted using a plant DNA extraction kit (TIANGEN Biotech, Beijing, China) and was now stored in the cryogenic refrigerator at Hebei Forest City Construction Technology Innovation Center, Shijiazhuang, China.
The Illumina HiSeq PE150 double-end sequencing strategy was used to build the library based on clean data at Beijing Medical Technology Co., Ltd. Then, GetOrganelle (https://github.com/Kinggerm/GetOrganelle) was used to assemble the plastid genome sequence. Finally, the GeSeq (Tillich et al. Citation2017) was used to annotate the plastid genomes of U. bergmanniana (MT108139). The results showed that the chloroplast genome of U. bergmanniana was a typical circular double-stranded tetrad with a total length of 160,060 bp, including a large single-copy region of 88,161 bp, a small single copy region of 19,029 bp and a pair of inverted repeat regions of 26,435 bp, respectively. The total GC content of the genome is 35.52%. A total of 131 genes were identified, including 86 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Among them, six protein-coding genes, six tRNA genes, and four rRNA genes were duplicated in the reverse repeat region.
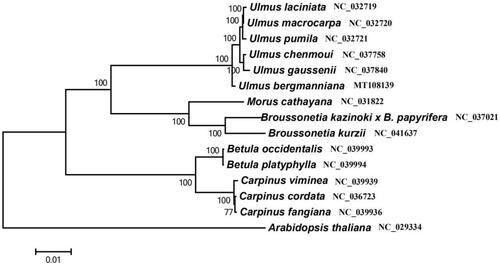
Phylogenetic tree using MEGA 7 (Kumar et al. Citation2016) was constructed by Maximum likelihood (ML) method and repeated 1000 times, based on the whole chloroplast genome sequences of U. bergmanniana and 14 other plants (including five Ulmaceae species, three Moraceae species, two Betula species, three Fagaceae species and a outgrope Arabidopsis thaliana) downloaded from NCBI database. The results showed that U. bergmanniana and the other five Ulmaceae plants were clustered into one branch, which was closely related to Moraceae. This result defines the phylogenetic position of U. bergmanniana at molecular level in Ulmaceae for the first time, and provides a theoretical basis for its molecular identification and resource development and utilizationt ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, under the accession no. MT108139. The associated BioProject number is PRJNA704466.
Additional information
Funding
References
- Chernik VV. 1981. Pseudomonomeric gynoecium of the Ulmaceae and Celtidaceae representatives. Bot Zhur. 66:958–962.
- Ebrahimi A, Antonides JD, Pinchot CC, Slavicek JM, Flower CE, Woeste KE. 2020. The complete chloroplast genome sequence of American elm (Ulmus americana) and comparative genomics of related species. Tree Genet Genomes. 17:5.
- Gu C, Ma L, Wu Z, Chen K, Wang Y. 2019. Comparative analyses of chloroplast genomes from 22 Lythraceae species: inferences for phylogenetic relationships and genome evolution within Myrtales. BMC Plant Biol. 19(1):281.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Palmer JD. 1985. Comparative organization of chloroplast genomes. Annu Rev Genet. 19(1):325–354.
- Shuxiang F, Yichao L, Shufang Y, Huang X, Yinran H. 2020. The complete chloroplast genome sequence of Ulmus lanceaefolia (Ulmaceae). Mitochondrial DNA B. 5(2):1605–1606.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Tobe H, Takaso T. 1990. Trichome micromorphology and evolution in Celtidaceae and Ulmaceae(Urticales). Amer J Bot. 77:2.
- Yinran H, Xiaoxu H, Shuxiang F, Shufang Y, Yongtan L, Yichao L. 2020. The complete chloroplast genome sequence of Ulmus elongata (Ulmaceae). Mitochondrial DNA B Resour. 5(1):792–793.
- Zavada M. 1983. Pollen morphology of Ulmaceae. Grana. 22(1):23–30.
- Zavada M, Crepet WL. 1981. Investigations of angiosperms from the middle Eocene of North American: flowers of the Celtidaceae. Amer. J Bot. 68(7):924–933.