Abstract
Tachysurus vachellii are commercially important edible fish due to delicious taste, little bone in muscle, and high nutritional value especially in Asia. The complete mitochondrial genome of Tachysurus vachellii has been sequenced. The mitochondrial genome is 16,529 bp in length, with the base composition of 31.6% A, 26.6% T, 26.9% C, and 14.9% G, containing two ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and a major non-coding control region (D-loop region). The gene order and orientation are similar with some typical fish species. The data will provide useful molecular information for phylogenetic studies concerning T. vachellii and its related species.
Tachysurus vachellii are commercially important edible fish due to delicious taste, little bone in muscle, and high nutritional value especially in Asia (Zhang et al. Citation2016a). However, due to overfishing, environmental pollution and other factors, the capture production of T. vachellii has been declining in the recent years (Zhang et al. Citation2016b). To benefit the sustainable utilization of T. vachellii fishery resource, it is necessary to determine the complete mitogenome sequence and make clear its phylogenetic relationships with closely related species. This information can also provide a theoretical basis for the studies on molecular systematics, stock evaluation, conservation genetics, and evolutionary adaptation mechanisms (Chak et al. Citation2020).
In the present study, the biological specimen was collected from Nansi Lake, north latitude 34°66″ and east longitude 117°20″, Jining city, China. They were preserved in 95% alcohol, which were stored in biology herbarium of Heze University under the voucher number: Zhangming214. All DNA were extracted using phenol–chloroform extraction methods and stored at −80 °C. The mitogenome was amplified by primers which were initially published (Zhang et al. Citation2015). Fragments generated from PCR amplification were sequenced using Sanger sequencing technology (Yuping et al. Citation2019). Sequenced fragments were assembled to create the complete mitogenome using CodonCode Aligner 5.1.5 (CodonCode Corporation, Dedham, MA), followed with sequence analysis, assembly, and visualization using SepMan and DNAMan (Wang et al. Citation2021). The complete mitogenome was annotated using the software of Sequin (version 15.10, http://www.ncbi.nlm.nih.gov/Sequin/). Transfer RNA genes and their potential cloverleaf structures were identified using tRNAscan-SE 1.21. All newly determined sequences from this study were deposited in GenBank database (accession number: MW288250).
The total length of T. vachellii mitochondrial DNA is 16,529 bp, which includes 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and two non-coding regions: a putative control region (D-loop region) and the origin of light-strand replication. All genes showed the typical gene arrangement conforming to the vertebrate consensus (Prosdocimi et al. Citation2012). The content is 31.6% for A, 26.6% for T, 26.9% for C, and 14.9% for G. A high A + T content indicates an obvious antiguanine bias commonly observed in fishes (Qiao et al. Citation2013). Except for eight tRNA (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) genes and ND6, most of the genes were encoded on the heavy strand (H-strand). All genes displayed the typical gene arrangement conforming to the vertebrate consensus (Chen et al. Citation2013). In the 13 protein-coding genes of mitochondrial genome, four overlaps are detected as shown in: ATP8-ATP6, ATP6-COXIII, ND4-ND4L, and ND5-ND6 sharing 10, 1, 7, and 4 nucleotides, respectively. The overlap of the ATPase genes appears to be common in most vertebrate mitochondrial genome (7–10 bp) (Broughton et al. Citation2001). Among the 22 tRNA genes, three overlaps are found, i.e. tRNAIle-tRNAGln, tRNAGln-tRNAMet, and tRNAThr-tRNAPro. There are also two overlaps between ND2 and tRNATrp and between ND3 and tRNAArg. The D-loop was 887 bp, flanked by tRNAPro and tRNAPhe genes.
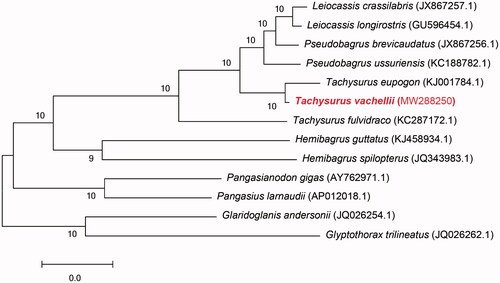
The maximum-likelihood tree by MEGA 7 based on 13 protein coding genes of the T. vachellii and the other 12 kinds of fish were constructed (Park et al. Citation2019). Seen from the phylogenetic tree (), T. vachellii have a closer relationship with Tachysurus eupogon. It is worth noting that a member of same genus, T. fulvidraco, was independent for one branch, which may be one of the reasons of mis-identification. Pseudobagrus, Leiocassis, Tachysurus clustered into Bagridae were the closest kinship groups. Compared with above groups, Hemibagrus was an early differentiation genus, followed by Pangasiidae. Above branches and Bagridae formed the sister group. Finally, they formed a sister group with Sisoridae.
Figure 1. Maximum-likelihood tree based on mitochondrial genome nucleotide sequences of the Tachysurus vachellii and the other 12 kinds of fish. Tachysurus vachellii (MW288250) in the position of the evolutionary tree. Numbers above branches are bootstrap values by 1000 replicates. The GenBank accession numbers of the sequences for the other 12 kinds of fish were used in the tree.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov, reference number MW288250.
Additional information
Funding
References
- Broughton RE, Milam JE, Roe BA. 2001. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11:1958–1967.
- Chak STC, Barden P, Baeza JA. 2020. The complete mitochondrial genome of the eusocial sponge-dwelling snapping shrimp Synalpheus microneptunus. Sci Rep. 10:47285.
- Chen Y, Cheng Q, Qiao H, Zhu Y, Chen W. 2013. The complete mitochondrial genome sequence of Rastrelliger kanagurta (Perciformes: Scombridae). Mitochondrial DNA. 24:114–116.
- Park J, Kim Y, Xi H. 2019. The complete mitochondrial genome sequence of Chinese minnow in Korea, Rhynchocypris oxycephalus (Sauvage and Dabry de Thiersant, 1874). Mitochondrial DNA B. 4:662–663.
- Prosdocimi F, Carvalho DCD, Beheregaray ALB. 2012. The complete mitochondrial genome of two recently derived species of the fish genus Nannoperca (Perciformes, Percichthyidae). Mol Biol Rep. 39:2767–2772.
- Qiao H, Cheng Q, Chen Y, Chen W, Zhu Y. 2013. The complete mitochondrial genome sequence of Coilia ectenes (Clupeiformes: Engraulidae). Mitochondrial DNA. 24:123–125.
- Wang Y, Du Y, Song X. 2021. Complete mitochondrial genome sequence of Anax parthenope (Odonata: Anisoptera: Aeshnidae) and phylogenetic analysis. Mitochondrial DNA B. 6(1):122–123.
- Yuping L, Xu S, Susup S, Lvsup T, Tao L. 2019. Characterization of the complete chloroplast genome sequence of Littledalea racemosa Keng (Poaceae: Bromeae). Mitochondrial DNA B. 13:33–35.
- Zhang G, Mao J, Liang F, Chen J, Zhao C, Yin S, Wang L, Tang Z, Chen S. 2016a. Modulated expression and enzymatic activities of Darkbarbel catfish, Pelteobagrus vachelli for oxidative stress induced by acute hypoxia and reoxygenation. Chemosphere. 151:271–279.
- Zhang G, Wang R, Mao J, Yin S, Chen S. 2015. The complete mitochondrial genome and phylogenic analysis of Pseudobagrus vachelli. Mitochondrial DNA. 27:1–2.
- Zhang G, Yin S, Mao J, Liang F, Zhao C, Li P, Zhou G, Chen S, Tang Z. 2016b. Integrated analysis of mRNA-seq and miRNA-seq in the liver of Pelteobagrus vachelli in response to hypoxia. Sci Rep. 6:22907.
