Abstract
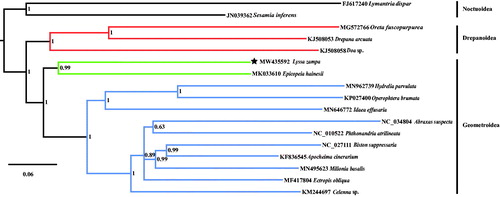
The complete mitochondrial genome (mitogenome) of Lyssa zampa was first reported. It is 15,314 bp in length (GenBank accession number: MW435592) and consists of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes. The nucleotide composition is A (41.5%), C (11.1%), G (7.4%), and T (40.0%). Based on the sequences of complete mitogenome from 12 geometroid species and three drepanoid species as ingroups, and two noctuoid species as outgroups, the phylogenetic tree was constructed. The results showed that the closest relationship between Uraniidae and Epicopeiidae was strongly supported by Bayesian posterior probabilities values of 0.99.
Lyssa zampa, which is broadly distributed from the Himalayas to Borneo and the Malay Peninsula, belongs to the family Uraniidae (Lepidoptera: Geometridae) (Nazari et al. Citation2016). Its larvae feed on Endospermum and other members of the rubber tree in the family Euphorbiaceae (Tokeshi and Yokoo Citation2007), and its genetic information is rarely known. Here, the complete mitochondrial genome (mitogenome) of L. zampa was first obtained and reported in the family Uraniidae, and the phylogenetic tree for understanding its phylogenetic position and relationships within the superfamily Geometridae was constructed. All the specimens and the genomic DNA in this study were deposited at the Insect Museum of Hunan Agricultural University, Changsha City, Hunan Province, China.
The specimens of L. zampa were collected on 21 July 2019 from Xiajinchang Town (23°N, 105°E, elevation of 1395 m) in Malipo County, Yunan Province, China. The complete genomic DNA was extracted following the methods in Wang et al. (Citation2019) from the legs of the adult (collection number: HAUHL033878) by using TaKaRa MiniBEST Universal Genomic DNA Extraction Kit Ver.5.0 (Shiga Prefecture, Kusatsu City, Japan), and was sequenced on the Illumina Hiseq platform with 150 bp paired-end reads at Novogene (Beijing, China). For Illumina TruSeq library, 6 Gb of clean data were obtained. The contigs and scaffolds of highly qualified sequences were determined using IDBA-1.1.3 (Peng et al. Citation2012; Chen et al. Citation2020). The mitogenome was annotated on the ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) and MITOS web server (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013).
The complete mitogenome of L. zampa is a circular DNA molecule of 15,314 bp in length (GenBank accession number: MW435592) and consists of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and an AT-rich region, in which 23 genes are transcribed on the J strand and the remaining 14 are transcribed on the N strand. The nucleotide composition is A (41.5%), C (11.1%), G (7.4%), T (40.0%), and the AT nucleotide content is 81.5%. There are 76 bp intergenic nucleotides that were dispersed in between 14 pairs of neighboring genes with their length varying from 1 to 24 bp. The length of the AT-rich region that located between rrnS and the trnM is 436 bp.
To construct the phylogenetic tree, the complete mitogenomes from 12 geometroid species and three drepanoid species as ingroups, and two noctuoid species as outgroups were used. Bayesian inference (BI) analysis was executed using MrBayes on XSEDE (3.2.6) (https://www.phylo.org/portal2/home.action), with the Markov chain Monte Carlo analysis run for 10,000,000 generations, sampled every 1000th generation and with a burn-in of 25%. The results are shown in . The family Uraniidae belongs to the superfamily Geometridae, which is consistent with Yang et al. (Citation2019). Meanwhile, the closest relationship between Uraniidae and Epicopeiidae was strongly supported by Bayesian posterior probabilities values of 0.99.
Acknowledgements
We thank Mr. Chang-Jin Yang for collecting the specimens.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication. The authors also are responsible for the content and writing of the paper.
Data availability statement
The mitogenome sequence data that support the finding of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm,nih.gov under the accession no. MW435592.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Juling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadier PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Chen L, Niklas W, Liao CQ, Wang CB, Ma FZ, Huang GH. 2020. Fourteen complete mitochondrial genomes of butterflies from the genus Lethe (Lepidoptera, Nymphalidae, Satyrinae) with mitogenome-based phylogenetic analysis. Genomics. 112:4435–4441.
- Nazari V, Schmidt BC, Prosser S, Hebert PDN. 2016. Century-old DNA barcodes reveal phylogenetic placement of the extinct Jamaican sunset moth, Urania sloanus Cramer (Lepidoptera: Uraniidae). PLoS One. 11:1–13.
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28:1420–1428.
- Tokeshi M, Yokoo M. 2007. New record of the tropical swallowtail moth Lyssa zampa (Butler) (Lepidoptera: Uraniidae) from mainland Japan. Entomol Sci. 10:103–106.
- Wang X, Chen ZM, Gu XS, Wang M, Huang GH, Zwick A. 2019. Phylogenetic relationships among Bombycidae s.l. (Lepidoptera) based on analyses of complete mitochondrial genomes. Syst Entomol. 44:490–498.
- Yang MS, Song L, Shi YX, Li JH, Zhang YL, Song N. 2019. The first mitochondrial genome of the family Epicopeiidae and higher-level phylogeny of Macroheterocera (Lepidoptera: Ditrysia). Int J Biol Macromol. 136:123–132.