Abstract
We determined the mitochondrial genome sequences of two snail mites, Riccardoella tokyoensis and R. reaumuri. The length of the entire mitogenome of these two species is 15,078 bp and 15,148 bp long, respectively. Both of them contain 13 proteins, two rRNAs, and 22 tRNAs for a total of 37 gene products. The gene order of Riccardoella is able to explain by a single rearrangement event from that of other Eupodina species; the whole region, including both rRNA genes and control region (CR), is inverted at the same position. The CR including a tandem repeat region in both of the mitogenomes of Riccardoella species.
So far, mites parasitizing land snails are known from three different families (Fain and Barker Citation2003). The first is the Eupodidae, mostly soil-borne, but only Eupodes voxencollinus Thor, 1934 has been found in the pallial cavity of bulimulid and helicid gastropods (Polaco and Mendl Citation1988) though this parasitism was probably accidental (Fain Citation2004). The second is the Trombiculidae, where larvae of Endotrombicula vanmoli (Vercammen-Grandjean & Benoit, 1971) have been reported to be deeply embedded in the soft integument (Fain Citation2004). The last family is Ereynetidae, and although unidentified species of the genus Boydaia also have been found on snails of which infection was also probably accidental (Polaco and Mendl Citation1988; Fain Citation2004), the most famous and globally distributed mite genus is Riccardoella Berlese, 1923 (Ereynetidae). This genus currently consists of eight species, and six of them have been recorded from the lungs of terrestrial gastropods (Turk and Phillips Citation1946; Fain and van Goethem Citation1986; Fain and Klompen Citation1990; André et al. Citation2004; Waki et al. Citation2018). The remaining two species were collected only from soils and thought to be free-living (Fain and van Goethem Citation1986; André et al. Citation2004). Among the mites belonging to the genus, Riccardoella limacum (Schrank, 1776) is known to parasitize several snails and sometimes cause a severe problem with edible snail farming (Baur and Baur Citation2005; Schüpbach and Baur Citation2008).
The phylogenetic relationships of Eupodides, including these snail mites, have been studied based on morphological information up to the early 2000s (Lindquist Citation1996; Andre and Fain Citation2000). Recent higher-level molecular phylogenetic studies indicated the uncertainly of the monophyly of Supercohort Eupodides. Eupodides was a polyphyletic group in Dabert et al. (Citation2016) and one of the superfamily Eriophyoidea was unstable among the markers in Klimov et al. (Citation2018). Waki et al. (Citation2018) used COI partial sequences to clarify the phylogenetic position of the genus Riccardoella among Eupodides. The snail mites were situated inside the superfamily Tydeoidea. However, the monophyly of the two superfamilies themselves and the families' relationships consisting of the superfamilies was unclear.
Thus, further molecular markers and more taxon sampling are urgent to elucidate phylogenetic relationships and genetic structure for revising taxonomy and species diversity in Ereynetidae mites. However, there was no mitogenome record for the superfamily Tydeoidae, and only five mitogenome sequences were reported from species of supercohort Eupodides. Hence, we choose two Riccardoella species for the representative of the superfamily and determined the whole mitogenome sequences by shotgun sequencing for both species.
Samples of Riccardoella tokyoensis were collected with host species (Tauphaedusa tau) at Rinshi no Mori Park, Tokyo (35.6243 N 139.7035 E). For R. reaumuri, host snails (Euhadra callizona) were collected at Nishizato, Shizuoka (35.1137 N 138.4168 E). Total DNA was extracted using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) and processed by QIAseq FX DNA Library kit (QIAGEN, Hilden, Germany). Paired-end sequencing (300 cycles) was conducted by the National Museum of Nature and Science, Tokyo on MiSeq, with inserts of ca. 50–200 bp, for a total of ca. four million reads. Assembly was performed using CLC Genomics Workbench ver. 12 (QIAGEN, Hilden, Germany) with the default setting. The ambiguous part of the contig was proofread by 3500 xL Genetic Analyzer (Thermo Fisher Scientific Co., Waltham, MA). Gene identification was made using MITOS web server (Bernt et al. Citation2013) and ARAGORN ver. 1.2.38 (Laslett and Canbäck Citation2004, 2008). Voucher specimens with extracted DNA were deposited at the National Museum of Nature and Science, Tokyo (NSMT-DNA 50369 and 50371).
The determined mitogenome length of Riccardoella tokyoensis Waki & Shimano, 2018 (GenBank/DDBJ/EMBL accession number LC601992) and R. reaumuri Fain and van Goethem, Citation1986 (LC601993) is 15,078 bp and 15,148 bp long, respectively. Both of them contain 13 proteins, two rRNAs, and 22 tRNAs for a total of 37 gene products. The overall A + T content of the R. tokyoensis and R. reaumuri mitochondrial genome is 79.9% and 81.6%, respectively, which is slightly higher than the ordinal range among Eupodina species (66.3–78.6%). In the mitogenome of R. tokyoensis and R. reaumuri, ATP8 starts with ATC/ATT codon, respectively. The gene order of Riccardoella is able to explain by a single rearrangement event from that of other Eupodina species. The whole region, including both rRNA genes and the control region (CR), is inverted at the same position. CR including tandem repeat region in both of the mitogenomes of Riccardoella species.
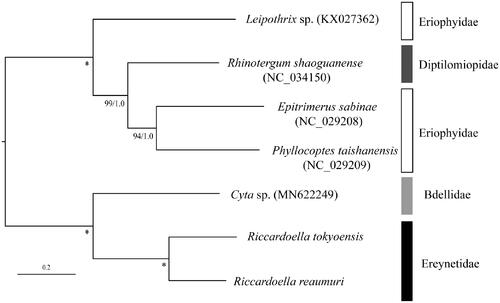
The maximum-likelihood (ML) phylogenetic analysis based on translated amino-acid sequences of 13 protein coding genes was conducted by RAxML-NG ver.1.0.1 (Kozlov et al. Citation2019) with bootstrap analyses of 1000 replicates. The phylogenetic tree also with posterior probability from Bayesian analyses (BAs) conducted by MrBayes 3.2.6 (Ronquist et al. Citation2012). In the tree, R. tokyoensis and R. reaumuri made a sister clade with species belonging to the family Bdellidae with high nodal support value (). The species of the family Eriophyidae formed a polyphyletic group, nested with Rhinotergum shaoguanense Xue, Song & Hong, 2009 (Diptilomiopidae). Although additional OTUs are needed, this mitogenome would be useful for reconstructing higher systematics of Eupodides mites.
Figure 1. Maximum-likelihood tree based on the concatenated nucleotide sequence of 13 protein-coding genes of Riccardoella tokyoensis (LC601992) and R. reaumuri (LC601993), five further Eupodides species. Accession numbers of the mitogenome sequences for each taxon used in the phylogenetic analysis are shown in parentheses. Nodal values are ML bootstrap support (BS) values and BA posterior probabilities (PPs). *100% BS and 1.0 PP. The scale bar indicates branch length in substitutions per site. PartitionFinder 2.1.1 (Lanfear et al. Citation2017) was used to determine the best partitioning scheme and the substitution model with branch lengths linked and a greedy search algorithm (Lanfear et al. Citation2012). The optimal partitioning strategy and evolutionary models consisted of thirteen genes data set for ML analyses were as follows; partition 1 (ND2 and ND3), partition 2 (CYTB, COII, and COIII), partition 3 (ATP6), partition 4 (ATP8), and partition 5 (ND6) with mtZOA + G+F; partition 6 (COI) with mtART + G+F; partition 7 (ND1, ND4, ND4L, and ND5) with mtZOA + I+G + F. For BA analysis, partition 1 (COI) and partition 2 (CYTB) with mtREV + G; partition 3 (rest of 11 PCGs) with GTR + I+G.
Acknowledgements
We express our gratitude toward the staff of Tokyo Metropolitan Government Bureau of Construction Tobu (Eastern) District Park Office and service center of Rinshi-no-mori Park, Tokyo for their kind support regarding this research.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information database (NCBI/GenBank) at https://www.ncbi.nlm.nih.gov/, accession numbers LC552026 and LC552027. Voucher specimens with extracted DNA were deposited at the Center for Molecular Biodiversity Research, National Museum of Nature and Science, Tokyo (Makoto Manabe; [email protected]) under the catalog numbers NSMT-DNA 50369 and 50371.
Additional information
Funding
References
- André HM, Ducarme X, Lebrun P. 2004. New ereynetid mites (Acari: Tydeoidea) from karstic areas: true association or sampling bias? J Cave Karst Stud. 66:81–88.
- Andre HM, Fain A. 2000. Phylogeny, ontogeny and adaptive radiation in the superfamily Tydeoidea (Acari: Actinedida), with a reappraisal of morphological characters. Zool J Linn Soc. 130(3):405–448.
- Baur A, Baur B. 2005. Interpopulation variation in the prevalence and intensity of parasitic mite infection in the land snail Arianta arbustorum. Invertebr Biol. 124(3):194–201.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Dabert M, Proctor H, Dabert J. 2016. Higher-level molecular phylogeny of the water mites (Acariformes: Prostigmata: Parasitengonina: Hydrachnidiae). Mol Phylogenet Evol. 101:75–90.
- Fain A. 2004. Mites (Acari) parasitic and predaceous in terrestrial gastropods. In: Barker GM, editor. Natural enemies of terrestrial molluscs. Wallingford, Oxfordshire, England: CABI Publishing.
- Fain A, Barker GM. 2003. A new genus and species of mite of the family Ereynetidae (Acari Prostigmata) from the pallial cavity of a New Zealand terrestrial gastropod (Athoracophoridae). Bull Soc R Belge D’Entomol. 139:233–238.
- Fain A, Klompen JSH. 1990. Riccardoella (Proriccadoella) triodopsis nov. spec. (Acari: Ereynetidae) from the USA. Acarologia. 31:187–190.
- Fain A, van Goethem JL. 1986. Les acariens du genre Riccardoella Berlese, 1923 parasites du poumon de mollusques gastéropodes pulmonés terrestres. Acarologia. 27:125–140.
- Klimov PB, OConnor BM, Chetverikov PE, Bolton SJ, Pepato AR, Mortazavi AL, Tolstikov AV, Bauchan GR, Ochoa R. 2018. Comprehensive phylogeny of acariform mites (Acariformes) provides insights on the origin of the four-legged mites (Eriophyoidea), a long branch. Mol Phylogenet Evol. 119:105–117.
- Kozlov A, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35:4453–4455.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Laslett D, Canbäck B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Lindquist EE. 1996. Phylogenetic relationships, 1.5.2. In: Lindquist EE, Bruin J, Sabelis MW, editors. Eriophyoid mites: their biology, natural enemies and control. Vol. 6. Amsterdam, The Netherlands: Elsevier; p. 301–327.
- Polaco OJ, Mendl W. 1988. Occurrence of mites in Mexican land snails. Nautilus. 102(3):129.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Schüpbach HU, Baur B. 2008. Parasitic mites influence fitness components of their host, the land snail Arianta arbustorum. Invertebr Biol. 127(3):350–356.
- Turk FA, Phillips S. 1946. A monograph of the slug mite—Riccardoella limacum (Schrank.). Proc Zool Soc Lond. 115:448–472.
- Waki T, Hiruta SF, Shimano S. 2018. A new species of the genus Riccardoella (Acari: Prostigmata: Ereynetidae) from the land snail Tauphaedusa tau (Gastropoda: Clausliidae) in Japan. Zootaxa. 4402(1):163–174.
