Abstract
Paeonia rockii subsp. taibaishanica (Paeoniaceae), one of the tree peony species, is endemic to the Qinling Mountains in central China. In this study, we characterized its whole plastid genome sequence using the Illumina sequencing platform. The complete plastid genome size of P. rockii subsp. taibaishanica is 153,368 bp in length, including a large single copy (LSC) region of 85,030 bp, a small single copy (SSC) region of 17,042 bp, and a pair of inverted repeats (IRs) of 25,648 bp. The genome contains 131 genes, including 83 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The GC contents in chloroplast genome, LSC region, SSC region, and IR region were 38.3%, 36.6%, 32.6%, and 43.1%, respectively. A total of 16 species are used to construct the phylogenetic tree of Paeoniaceae, the results showed that P. rockii subsp. taibaishanica is more closely related with congeneric Paeonia suffruticosa and Paeonia ostii, these species were clustered into a clade with high bootstrap support.
Paeonia rockii subsp. taibaishanica D. Y. Hong (Paeoniaceae), one of the tree peony species, is endemic to the Qinling Mountains in central China (Liu et al. Citation2019). The genus Paeonia, also known as the ‘King of Flowers’ in China, is an important source of traditional Chinese medicine (Bao et al. Citation2018). P. rockii subsp. taibaishanica have long been used to treat a range of cardiovascular and gynecological diseases (Liu et al. Citation2019). This species has been listed as endangered species in the IUCN Red List, and urgent management and conservation are required. In this study, we characterized the complete plastid genome sequence of P. rockii subsp. taibaishanica based on the Illumina pair-end sequencing data. The annotated chloroplast genome of P. rockii subsp. taibaishanica has been deposited into the GenBank with the accession number MW192444.
The fresh and healthy leaves of P. rockii subsp. taibaishanica were collected in Taibai county, Shaanxi Provincial National Nature Reserve (Baoji, China; N 34.062401, E107.700326; Alt.1564.41m). The dried tissue samples and voucher specimens (No. PRLZH2019022) were deposited in the Evolutionary Botany Group, Key Laboratory of Resource Biology and Biotechnology in Western China (Shaanxi, China). The total genomic DNA was extracted using the modified cetyl trimethyl ammonium bromide method (Doyle and Doyle Citation1987) and further assessed by agarose gel electrophoresis to check the quality of DNA. Then, the DNAs were subjected to Illumina sample preparation. We constructed a pair-end (PE) library with 350 bp insert size fragments using TruSeq DNA sample preparation kits (Sangon, Shanghai, China). The high throughput DNA molecular sequencing was indexed by the Illumina Hiseq 2500 technology platform. The obtained raw reads were trimmed using the program NGSQC Toolkit_version 2.3.3 with the default statistical genetic parameters (Patel and Jain Citation2012). After dislodged the low-quality DNA sequence reads, the clean DNA reads were assembled using the software MIRA version 4.0.2 (Chevreux et al. Citation2004) and MITObim version 1.8 (Hahn et al. Citation2013) with the complete plastid genome of congeneric P. rockii (NC037772) as the reference DNA sequences. The genome P. rockii subsp. taibaishanica was annotated by the programs PGA (Qu et al. Citation2019) and DOGAM (Wyman et al. Citation2014) (http://dogma.ccbb.utexas.edu/).
The complete plastid genome size of P. rockii subsp. taibaishanica is 153,368 bp in length, including a large single copy (LSC) region of 85,030 bp, a small single copy (SSC) region of 17,042 bp, and a pair of inverted repeats (IRs) of 25,648 bp. The genome contains 131 genes, including 83 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. At the same time, the three genes rpl22, rps18, and ycf1 were found to be pseudogenes. The GC content in plastid genome, LSC region, SSC region, and IR region were 38.3%, 36.6%, 32.6%, and 43.1%, respectively. Meanwhile, a total of 15 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rps16, rpoC1, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron, and three genes (ycf3, rps12, and clpP) contained two introns.
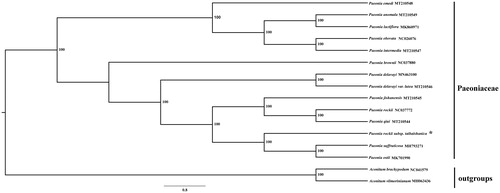
A total of 14 species from the genus Paeonia were used to construct the phylogenetic tree with two Aconitum species as outgroups. All of the 16 plastid sequences were aligned using the software MAFFT (Katoh and Standley Citation2013) with the default genetic parameters. The phylogenetic analysis was conducted using the program RAxML (Stamatakis Citation2006) with 1000 bootstrap replicates. The results showed that P. rockii subsp. taibaishanica was closely related to the two congeneric species Paeonia suffruticosa and Paeonia ostii ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the results of this work are openly available in GenBank at https://www.ncbi.nlm.nih.gov, reference number MW192444.
Additional information
Funding
References
- Bao Y, Qu Y, Li J, Li Y, Ren X, Maffucci K, Li R, Wang Z, Zeng R. 2018. In vitro and in vivo antioxidant activities of the flowers and leaves from Paeonia rockii and identification of their antioxidant constituents by UHPLC-ESI-HRMS via pre-column DPPH reaction. Molecules. 23(2):392.
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14(6):1147–1159.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Boil Evol. 30(4):772–780.
- Liu N, Cheng F, Zhong Y, Guo X. 2019. Comparative transcriptome and coexpression network analysis of carpel quantitative variation in Paeonia rockii. BMC Genomics. 20(1):683.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1746–4811.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2014. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.