Abstract
Pennisetum flaccidum Grisebach is a typical high-quality forage and adrought-tolerant grass. In this study, we firstly reported the complete chloroplast (cp) genome of P. flaccidum, which was 138,336 bp in length, including a pair of inverted repeats (IR: 22,293 bp), a large single copy (LSC: 81,329 bp), and a small single copy (SSC: 12,421 bp) region. A total of 131 genes were annotated, containing seven rRNA genes, 38 tRNA genes, and 86 protein-coding genes. The GC content of the cp genome was 38.63%. The maximum-likelihood (ML) phylogenetic tree indicated that P. flaccidum was closely related to P. cetaceum in Poaceae.
Pennisetum flaccidum Grisebach is a plant species of the genus of Pennisetum in the family of Poaceae and widely distributes in northern China with high quality and excellent palatability (Joshi and Ludri Citation1967; Yu et al. Citation2009). P. flaccidum has strong drought tolerance and can reproduce and grow multiply on sandy land or dunes. It is not only one of the herbaceous species with rich nutrient but also a pioneer species for sand fixation (Editorial Board of Forage flora of China Citation1987). The analysis of complete chloroplast (cp) genomes has a broad prospect in understanding plant genetic diversity, plant distribution, and phylogenetic evolution (Yang et al. Citation2019). In this study, we employed Illumina Hiseq to determine and assemble the complete cp genome of P. flaccidum, to provide novelty and information on the understanding of its phylogenetic status.
The fresh leaves of P. flaccidum were collected from Southwestern Mu Us Sandy Land, China (106°24′E, 37°57′N) and dried with silica gel. A specimen (R02) was deposited in the Herbarium of School of Agriculture, Ningxia University (https://nxy.nxu.edu.cn/, Wangsuo Liu [email protected]) under the voucher number 6401812020-LW001. The modified CTAB method was used to extract the whole-genomic DNA (Stefanova et al. Citation2013). The Illumina Hiseq PE150 sequencing was conducted by Biomarker Technologies Co., Ltd. (Qingdao, China). After removal and filtration of low-quality reads, in total, 5 Gb clean reads (the average Q30 was 92.14%) were obtained. The qualified reads were assembled using SPAdes version 3.11.0 software
Q5 (Bankevich et al. Citation2012). The genome annotation was performed by GeSeq (Tillich et al. Citation2017). Draw annotation map was used by OGDRAW (Greiner et al. Citation2019) in web service (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). The complete cp genome
Q3 sequence and annotation of P. flaccidum were submitted to GenBank (accession No. MW442087).
The borders of IR, LSC, and SSC regions were identified using an online tool, IRscope (Amiryousefi et al. Citation2018). The cp genome of P. flaccidum was 138,336 bp in length and with four typical regions, containing two inverted repeats (IRa and IRb: 22,293 bp), a large single copy (LSC: 81,329 bp), and a small single copy (SSC: 12,421 bp) region. A total of 131 genes were annotated, including seven rRNA genes, 38 tRNA genes, and 86 protein-coding genes. The GC content of the cp genome was 38.63%. In these genes, there were 93 unique genes and 19 were duplicated in the IR regions. The 19 duplicates including eight protein-coding genes (rps19, rpl2, rpl23, ycf2, ndhB, rps7, rps12, and rps15), eight tRNA genes (trnH-GUG, trnl-CAU, trnL-CAA, trnV-GAC, trnl-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and three rRNA genes (rrn4.5, rrn5, and rrn16).
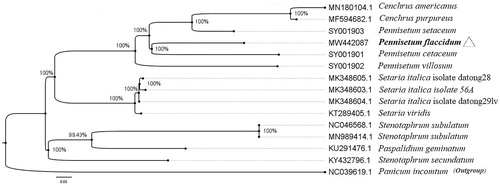
In order to clarify the phylogenetic location of P. flaccidum, the cp genome sequence of 12 Poaceae was downloaded from NCBI and added three whole cp genome sequences of Pennisetum (SY001901, SY001902, and SY001903) at https://lcgdb.wordpress.com/. All cp genomes were aligned by MAFFT version 7.037 (Katoh and Standley Citation2013). Maximum-likelihood (ML) tree was constructed by PhyML-3.1(GTR) (Guindon et al. Citation2010) with 1000 bootstrap replications, and the diagram was processed by FigTree version 1.4.4 as shown in the . The result showed that P. flaccidum closely clustered with P. cetaceum. Our study provides a reference for the phylogenetic and taxonomic status of Pennisetum in Poaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI database at https://www.ncbi.nlm.nih.gov/, reference number MW442087. The associated BioProject, Biosample, and SRA numbers are PRJNA689090, SAMN17199738, and SRS7976986, respectively.
Additional information
Funding
References
- Amiryousefi A, Hyvonen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34(17):3030–3031.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Editorial Board of Forage flora of China. 1987. Forage flora of China. Vol. 1. Beijing, China: Agricultural Press; p. 139–140.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucl Acid Res. 47(W1):W59–W64.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Joshi DC, Ludri RS. 1967. The chemical composition and nutritive value of Buffle (Pennisetum flaccidum, Griseb) and Panicum kabulabula grasses. Indian Vet J. 44(9):800–804.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Stefanova P, Taseva M, Georgieva T, Gotcheva V, Angelov A. 2013. A modified CTAB method for DNA extraction from soybean and meat products. Biotechnol Biotec Eq. 27(3):3803–3810.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucl Acid Res. 45(W1):W6–W11.
- Yang C, Wu X, Guo X, Bao P, Chu M, Zhou X, Liang Z, Ding X, Yan P. 2019. The characterization of the complete chloroplast genome of wild Elymus nutans. Mitochondrial DNA Part B. 4(1):925–926.
- Yu YW, Nan ZB, Hou FJ, Matthew C. 2009. Response of Stipa bungeana and Pennisetum flaccidum to urine of sheep in steppe grassland of north-western China. Grass Forage Sci. 64(4):395–400.