Abstract
We report the complete mitochondrial genome (mtDNA) of Pareas formosensis (Squamata: Colubridae). This circular mtDNA is 17,703 bp in size and consists of 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs, and 2 non-coding sequence (D-loop). The total of mtDNA was composition of 57.26% A + T and 42.74% G + C (T: 25.21%, C: 28.84%, A: 32.05%, G: 13.90%). The phylogenetic analysis revealed that P. formosensis formed a clade with other species of Pareas. This mtDNA sequence of P. formosensis provides useful data for studying the population genetics and phylogeography of Colubridae.
The Pareatidae was divided into three genera: Aplopeltura, Asthenodipsas and Pareas (Guo and Zhang Citation2015). Pareas is the largest genus, which contains 20 species (Bhosale et al. Citation2020). Pareas formosensis (Van Denburgh 1909) is widespread in South China (Anhui, Jiangsu, Jiangxi, Zhejiang, Fujian, Sichuan, Guizhou, Yunnan, Guangxi, Guangdong, Taiwan) (Zhao Citation2006). In this genus, the complete mitochondrial genome (mtDNA) was described only in P. boulengeri (Huang et al. Citation2020). Here, we determined the mtDNA of P. formosensis in order to provide useful data for studying the population genetics and phylogeography of Colubridae.
The specimen (species voucher: LSU2020MLTWDT01) was collected in Baiyunshan, from Lishui, Zhejiang Province, China (N28.498097°, E119.920808°), then placed in 90% ethanol and stored in Laboratory of Amphibian Diversity Investigation (Contact person: Li Ma, E-mail: [email protected]) at Lishui University, China. The total DNA was extracted from muscles using the EasyPure genomic DNA kit (Trans Gen Biotech Co., Beijing, China) from the muscle tissue of P. formosensis, and then was sequenced by a sequencing company (Novogene Bioinformatics Technology Co. Ltd., Tianjin, China). We used MITOS web server to carry out and annotate the gene sequence (Matthias et al. Citation2013). The mtDNA of P. formosensis is a closed-circular molecule of 17,703 bp in length. The complete mtDNA of P. formosensis (Genbank accession No MW531674) contains 13 protein-coding genes (PCGs), 22 tRNAs and 2 rRNAs, two D-loop and a L-chain replication-initiating non-coding region (NCR). The gene sequence of P. formosensis is approximately the same as P. boulengeri (Huang et al. Citation2020). 13 PCGs include NAD1-6, COX1-3, ATP6, ATP8, NAD4L, and CYTB, among which NAD5 is the longest (1,788bp) and ATP8 is the shortest (162 bp). The ND6 subunit gene and eight tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAPro, and tRNAGlu) were encoded on the L-strand, whereas the other genes were encoded on the H-strand. All PCGS start with an ATG codon except ND3 begins with ATA codon, and COX1 starts with GTG. Six genes (NAD1, NAD3-4, CYTB, and COX2-3) end with incomplete stop codons (T–/TA-), and four genes (NAD5, ATP6, ATP8, and NAD4L, and CYTB) end with TAA, COX1 and NAD6 end with AGA and NAD2 end with TAG. The overall base composition for mtDNA sequence of P. formosensis is as follows: T (25.21%), C (28.84%), A (32.05%) and G (13.90%).
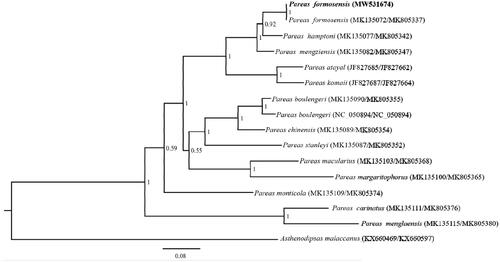
Based on the CYTB (1086 bp) and NAD4 (663 bp) with Asthenodipsas malaccanus as outgroup by using Bayesian inference (BI) methods in MrBayes v3.2.2 (), phylogenetic analyses of P. formosensis and other 14 species of Pareas were conducted. We used MrModelTest 2.3 to find the best-fit substitution model (GTR + I + G) (Nylander Citation2004). Phylogenetic analysis revealed that P. formosensis was more closely related to P. hampton than other species. P. carinatus and P. menglaensis has the furthest relationship with P. formosensis. The result is consistent with previously reported (You et al. Citation2015; Wang et al. Citation2020). Essentially, it is useful for subsequent research about the population genetics and phylogeography of Colubridae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitogenome data supporting this study are openly available in GenBank at [https://www.ncbi.nlm.nih.gov/nuccore/ MW531674]. Reference number [Accession number: MW531674]. BioSample and SRA accession numbers are [https://www.ncbi.nlm.nih.gov/biosample/SAMN17394016], [https://www.ncbi.nlm.nih.gov/sra/SRR13495178], respectively.
Additional information
Funding
References
- Bhosale H, Phansalkar P, Sawant M, Gowande G, Patel H, Mirza ZA. 2020. A new species of snail eating snakes of the genus Pareas Wagler, 1830 (Reptilia: Serpentes) from eastern Himalayas, India. Eur J Taxon. 729:54–73.
- Guo YH, Zhang QL. 2015. Review of systematics on the Asian snail-eating snakes. Chinese Journal of Zoology. 2015, 50(1): 153–158.
- Huang RY, Peng LF, Yang DC, Yong Z, Huang S. 2020. Mitochondrial genome of the Boulenger's Slugeating snake Pareas boulengeri (Serpentes: Pareidae). Mitochondrial DNA Part B. 5(3):3179–3180.
- Matthias B, Alexander D, Frank J, Fabian E, Catherine F, Guido F, Joern P, Martin M, Peter FS. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet E. 69(2):313–319.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Wang P, Che J, Liu Q, Li K, Jin JQ, Jiang K, Shi L, Guo P. 2020. A revised taxonomy of Asian snail-eating snakes Pareas (Squamata, Pareidae): evidence from morphological comparison and molecular phylogeny. ZooKeys. 939:45–64.
- You CW, Poyarkov NA, Lin SM. 2015. Diversity of the snail-eating snakes Pareas (Serpentes, Pareatidae) from Taiwan. Zool Scr. 44(4):349–361.
- Zhao EM. 2006. Snakes of China. Hefei (China): Anhui Sciences and Technology Publishing House. (In Chinese).