Abstract
Epimedium flavum Stearn, which belongs to Berberidaceae, is mainly distributed in the Sichuan province of China. In this study, the complete chloroplast genome of E. flavum was reported for the first time. The whole genome of E. flavum was 159,134 bp in length, and revealed a typical quadripartite structure, including two copies of an inverted repeat (IR) region of 27,735 bp separating a large single-copy region (LSC, 86,576 bp) and a small single-copy region (SSC, 17, 088 bp). The chloroplast genome contained 112 unique genes, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis showed that E. flavum of series Davidianae was firstly clustered with E. brevicornu of ser. Brachyerae.
Epimedium L. contains about 62 species, which are discontinuously distributed in the Mediterranean region and eastern Asia (Ying et al. Citation2011; Zhang et al. Citation2020). As the diversity center of Epimedium, China possesses about 52 species of the genus and has used Epimedium plants as traditional medicine ‘Herba Epimedii’ for more than 2000 years (Zhao et al. Citation2012). Modern pharmacological studies show that Herba Epimedii can be widely used against sexual dysfunction, osteoporosis, and inflammation (Ma et al. Citation2020). However, Epimedium has abundant variations in morphology and medicinal ingredients, which greatly increases the difficulty of species identification (Zhang et al. Citation2016). In recent years, with the development of modern molecular techniques, the chloroplast genome has been widely used to study the genetic diversity and phylogenetic relationship in plant species (Yang et al. Citation2019). In this study, we sequenced the complete chloroplast genome of E. flavum, aiming to provide more valuable information about the phylogenetic relationships of Epimedium species.
In this study, the E. flavum samples were collected from the Tianquan County, Sichuan, China (102°16′E, 30°06′N), and a Yanjun Zhang 566 (HIB) voucher was deposited at the Herbarium of Wuhan Botanical Garden, Chinese Academy of Sciences (HIB). The genomic DNA was extracted from the fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987). Genome sequencing was sequenced using Illumina Novaseq PE150, and 150 bp paired-end reads were generated. The filtered reads were assembled using the program GetOrganelle v1.7.2a with E. acuminatum chloroplast genome (GenBank accession number: NC_029941) as a reference (Jin et al. Citation2020). The chloroplast genome sequence was annotated through the online program CPGAVAS 2, followed by manual correction (Shi et al. Citation2019). The chloroplast genome sequence of Epimedium flavum was submitted to the NCBI database with an accession number (MW467895).
The genome sequence of E. flavum was 159,134 bp in length, and displayed a typical quadripartite structure, including two copies of an inverted repeat (IR) region of 27,735 bp separating a large single-copy region (LSC, 86,576 bp) and a small single-copy region (SSC, 17,088 bp). The overall GC content of chloroplast genomes was 38.81%, while the GC content in LSC IR and SSC regions was 37.29%, 43.03%, and 32.79%, respectively. The chloroplast genome of E. flavum contained 112 unique genes, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Among these genes, six tRNA genes and nine protein-coding genes contained one intron, while three genes contained a pair of introns.
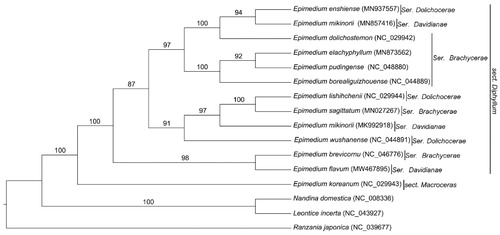
To identify the phylogenetic relationship of E. flavum, 15 complete chloroplast genomes of Berberidaceae species were downloaded from the NCBI GenBank database. The sequences were aligned using MAFFT v7 (Katoh et al. Citation2017), and the maximum-likelihood tree was constructed with 1000 bootstrap replicates using raxmlGUI1.5b (v8.2.10) (Silvestro and Michalak Citation2012; Katoh et al. Citation2017) (). Phylogenetic analysis showed that E. koreanmum of sect. Macroceras was separated from all the twelve species of the sect. Diphyllum of Epimedium. E. flavum of series Davidianae was firstly clustered with E. brevicornu of ser. Brachyerae, both of which are distributed in the Sichuan Basin, next to the eastern Tibet Plateau. The complete chloroplast genome of E. flavum contributes to the phylogenetic and evolutionary analysis of Epimedium species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW467895. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA714816, SRR13983522, and SAMN18318609, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):242.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 4:1–7.
- Ma DL, Zhang L, Li L. 2020. Anti-inflammatory effects and underlying mechanisms of Epimedium extracts. Prog Biochem Biophys. 47(8):685–699.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyser. Nucleic Acids Res. 47:65–73.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Ying TS, Boufford DE, Brach AR. 2011. Epimedium L. Vol. 19. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Beijing (China): Science Press; St. Louis: Missouri Botanical Garden Press; p. 787–799.
- Yang Y, Zhang Y, Chen Y, Gul J, Zhang J, Liu Q, Chen Q. 2019. Complete chloroplast genome sequence of the mangrove species Kandelia obovata and comparative analyses with related species. PeerJ. 7:e7713.
- Zhang Y, Du L, Liu A, Chen J, Wu L, Hu W, Zhang W, Kim K, Lee SC, Yang TJ, et al. 2016. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 7:306.
- Zhang YJ, Huang RQ, Wu L, Wang Y, Jin T, Liang Q. 2020. The complete chloroplast genome of Epimedium brevicornu Maxim (Berberidaceae), a traditional Chinese medicine herb. Mitochondrial DNA Part B. 5(1):588–590.
- Zhao Y, Song H, Fei J, Liang Y, Zhang B, Liu Q, Wang J, Hu P. 2012. The effects of Chinese yam-epimedium mixture on respiratory function and quality of life in patients with chronic obstructive pulmonary disease. J Tradit Chin Med. 32(2):203–207.