Abstract
The Pegu Rice Frog, Microhyla berdmorei is distributed across ten Asian countries. However, the DNA barcoding information (COI gene) is restricted to only Southeast Asian countries. Here, we sampled a specimen of M. berdmorei in Mizoram state, northeast India to allow the genetic diversity of the species across its range. We generated both COI and 16S ribosomal RNA sequences of the studied species to check the population genetic diversity. The Bayesian analyses clearly discriminate M. berdmorei from its sister species Microhyla pulchra. The present datasets of M. berdmorei also revealed 11 and 19 haplotypes with high uncorrected pairwise genetic distances in COI (3.8–11.8%) and 16S rRNA (0–4.6%) gene, respectively. Owing to the high intra-species genetic distances and different haplotypes with sufficient mutational steps in both mitochondrial genes, this study affirms the existence of M. berdmorei species complex or cryptic diversity within its range distribution in South and Southeast Asia.
Introduction
The genus Microhyla (family: Microhylidae) contains 50 species around the world; among them, 14 species are reported from India (Frost Citation2020). This microhylid genus is small-sized terrestrial frogs distributed in the Oriental biogeographic region (Poyarkov et al. Citation2014, Citation2018a, Citation2018b, Citation2019). Northeast India shares two biodiversity hotspots and is regarded as one of the prolific regions (with 153 known species) for global amphibian diversity (Biju et al. Citation2019). A total of five Microhyla species, M. berdmorei, M. butleri, M. eos, M. mukhlesuri, and M. mymensinghensis are reported from this region. Among them, M. berdmorei was described early and is distributed in Bangladesh, Cambodia, China, India, Indonesia, Laos, Malaysia, Myanmar, Thailand, and Vietnam (Hasan et al. Citation2014; Biju et al. Citation2019). This species generally inhabits moist evergreen forest adjacent to hilly regions and is often found near streams (van Dijk et al. Citation2004). The species is assessed to be ‘Least Concern’ by the International Union for Conservation of Nature Red List of Threatened Species (IUCN Citation2021); however, immense deforestation and other anthropogenic threats may decline the local population of M. berdmorei throughout its range distribution. Although, the recent discovery of M. eos from northeast India has increased the checklist of Microhyla, this genus contributed to present persistent taxonomic challenges (Gorin et al. Citation2020). Nevertheless, high homoplasy and cryptic diversity often obscured accurate diversity estimate of microhylids (Rakotoarison et al. Citation2017).
The integrated approach combining morphology, molecular, and bioacoustics is evidenced to be the best exercise to illuminate the anuran systematics (Frost et al. Citation2006; Van Bocxlaer et al. Citation2006; Köhler et al. Citation2017). The species diversity, patterns of distribution, phylogenetic relationships, biogeographic origin, and possible routes of colonization of microhylids, including the genus Microhyla have been repeatedly discussed in several studies (Matsui et al. Citation2011; De Sá et al. Citation2012; Peloso et al. Citation2016; Seshadri et al. Citation2016; Feng et al. Citation2017; Tu et al. Citation2018; Vineeth et al. Citation2018; Garg et al. Citation2019; Garg and Biju Citation2019; Poyarkov et al. Citation2019; Gorin et al. Citation2020). However, most of the studies deal with mitochondrial ribosomal RNA genes (12S rRNA and 16S rRNA) to define their existing biological questions (Vences et al. Citation2005; Vieites et al. Citation2009). As a result, the well-recognized and widely used DNA barcoding (mitochondrial Cytochrome Oxidase subunit I gene) information (Hebert et al. Citation2003a, Citation2003b; Biju et al. Citation2014; Chambers and Hebert Citation2016) of Microhyla is scanty in the global database, especially from northeast India. As of now, a few mtCOI sequences of M. berdmorei have been generated from Southeast Asian countries to improve the biodiversity assessment from different life stages and fill the Barcode Index Numbers (BINs) of the Barcode of Life Data System (BOLD) (Grosjean et al. Citation2015; Mulcahy et al. Citation2018). Nevertheless, most microhylid studies were based on stable species systematics, allowing assessment of their population genetic diversity. Considering the inadequacy of DNA sequence information of M. berdmorei, the present study intended to generate both mitochondrial COI and 16S rRNA sequences from northeast India, to aid our understanding of genetic diversity in this species. We used both phylogeny and haplotype analyses to estimate the population structure of this microhylid frog from its known range distribution. This primary genetic information will assist rapid and reliable estimates of population genetic diversity.
Materials and methods
An adult specimen of a microhylid frog was collected from Reiek Community Reserve Forest (23.75 N 92.67 E, 1190 m ASL), Mamit district, Mizoram state in northeast India (). The exploration was accomplished after acquiring the prior permission (No.A.33011/2/99-CWLW/225) from the Chief Wildlife Warden, Govt. of Mizoram, India. The collected specimen was identified as M. berdmorei based on the below morphological characters (). A small frog with smooth skin; dorsally varying from light to dark olive, sometimes with dark brownish spots or marbling scattered on dorsal surface of body; characteristic dark sheds present dorsally in between the eyes, running up to trunk region; few minute light brown spots present on lateral parts of hind limbs as well as on flanks; both throat and chest mottled with dark brown; limbs with faint cross bands; snout-vent length (SVL) of 28.6 mm to 35.7 mm. It can be diagnosed by having characteristic features like, head much broader than length, slightly depressed; snout pointed, a little longer than eyes, projecting a little beyond lower jaw; canthus rostralis prominent; nostrils a little closer to tip of snout than to eyes; tympanum hidden; vomerine teeth absent; forelimbs moderately long; fingers slender, free with rounded tips; length of fingers 1 < 2< 4 < 3, the third finger much longer than snout; subarticular tubercles large and prominent; hindlimbs very long; tibiotarsal articulation reaching beyond tip of snout; heels strongly overlapping when hind limbs folded at right angle to body; tibia 3½ to 4 times as long as broad, more than ⅔ the length of snout to vent; tips of toes swollen into rounded tips which are slightly larger than fingertips; digits with a median groove on the upper surface of the tip separating the upper surface from the lower ones; toes fully webbed; inner metatarsal tubercle prominent and oval; a small rounded outer metatarsal tubercle present (Lalremsanga Citation2011). The specimen was vouchered (MZMU 1613) in the museum of the Department of Zoology, Mizoram University, India. The muscle tissue was aseptically collected from the hind limb and preserved in 70% molecular grade ethanol.
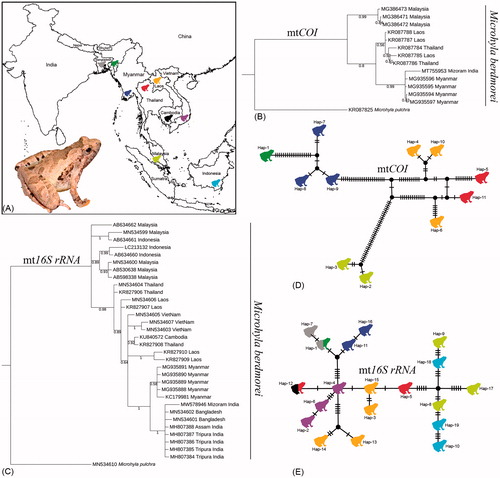
Figure 1. (A) Map showing the collection localities of Microhyla berdmorei and live individual of M. berdmorei collected from northeast India. (B, C) Bayesian phylogenies based on COI and 16S rRNA genes inferred the clustering pattern of M. berdmorei sampled from different geographical locations in South and Southeast Asian countries. (D, E) The TCS haplotype networks based on COI and 16S rRNA genes showed haplotype network of M. berdmorei collected from South and Southeast Asian countries. The estimated haplotypes are shown in different colored frog icons as represented in the collection locality map.
Total genomic DNA was extracted by using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) with standard manufacturer’s protocols. The genomic DNA was stored at −30 °C in the Center for DNA Taxonomy laboratory, Zoological Survey of India, Kolkata. The published primer pairs were used to amplify the partial fragment of mitochondrial COI and 16S rRNA genes on a Veriti® Thermal Cycler (Applied Biosystems, Foster City, CA) with standard thermal profiles (Palumbi Citation1996; Rassmann Citation1997; Ward et al. Citation2005). The 25 µl PCR mixture contains 10 pmol of each primer, 100 ng of DNA template, 1 × PCR buffer, 1.0–1.5 mM of MgCl2, 0.25 mM of each dNTPs, and 1 U of Taq polymerase (Takara BIO Inc., Japan). The amplified PCR products were visualized in 1% agarose gel containing Ethidium bromide (10 mg/ml). The PCR products were further purified by using the QIAquickR Gel extraction Kit (Qiagen, Valencia, CA). The cycle sequencing was performed with the purified PCR products (15 ng), BigDye®Terminator ver. 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA), and 3.2 picomoles of the same PCR primer pairs. The thermal profile of the cycle sequencing was set as 96 °C for 1 min, followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s, and a final extension at 60 °C for 1 min 15 s on the same thermal cycler. The cycle sequencing products were cleaned by BigDye X-Terminator Kit (Applied Bio systems, Foster City, CA), and Sanger sequencing was performed on 48 capillary ABI 3730 Genetic analyzer.
The generated bi-directional sequences were checked through SeqScanner V1.0 (Applied Biosystems Inc., CA) and nucleotide BLAST search platform (https://blast.ncbi.nlm.nih.gov/). The consensus sequences were submitted to the GenBank to acquire the accession numbers. The publicly available 12 COI sequences and 31 16S rRNA sequences of M. berdmorei were acquired from GenBank database. The sequence of closest species, Microhyla pulchra (COI: KR087825 and 16S rRNA: MN534610) were also acquired from GenBank and used as an outgroup in the Bayesian analysis (BA). The final datasets (16S rRNA: 564 bp and COI: 645 bp) were aligned separately through ClustalX (Thompson et al. Citation1997) and uncorrected pairwise (p) genetic distances were calculated through the MEGAX program with complete deletion of gap/missing data treatment setting (Kumar et al. Citation2018). The BA phylogenies were constructed in Mr. Bayes 3.1.2 (Ronquist and Huelsenbeck Citation2003) after estimating the best fit models (16S rRNA: T92 + G with lowest BIC score 2510.578 and COI: HKY + G with lowest BIC score 3530.804) through Mr.Modeltest v2 (Nylander Citation2004). The Markov chain Monte Carlo (one cold and three hot chains) was run for 1,000,000 generations with 25% burn-in and trees saving at each 100 generation, until the standard deviation of split frequencies reached 0.01 and the potential scale reduction factor for all parameters bordered on 1.0. Both phylogenies were illustrated in the iTOL tool for better representation (https://itol.embl. de/) (Letunic and Bork Citation2007). The haplotype networks for both genes were built to elucidate the haplotype diversity within the different population of M. berdmorei. The numbers of haplotypes, haplotype diversity (Hd), and the number of polymorphic sites were estimated through DnaSP v6 (Rozas et al. Citation2017). The haplotype networks was built through PopART (http://popart.otago.ac.nz) (Leigh and Bryant Citation2015) using the standard Templeton, Crandall and Sing (TCS) method (Clement et al. Citation2000).
Results and discussion
The plethora of biodiversity in northeast India has gained worldwide attention due to its unique biogeography, representing two global biodiversity hotspots (Eastern Himalaya and Indo-Burma) (Tordoff et al. Citation2011). Remarkably, the majority of the faunal composition of this region shares their range distribution with neighboring South and Southeast Asian countries due to the transcontinental dispersion (De Bruyn et al. Citation2014; Das et al. Citation2019). To date, this region supports 153 anuran species, among them 51 were described since 2001 (Kamei et al. Citation2012; Biju et al. Citation2019). Owing to the lack of accessibility exacerbated by difficult terrains and low-lying river basins, communication gap with the variety of ethnic communities and their indigenous languages, the exploration of anurans has been limited. Besides the few opportunistic sampling and subsequent taxonomic research, the large-scale study dealing with the integrated approach is meagerly attempted to explore the anuran diversity in this region.
The collected frog specimen was identified as M. berdmorei based on the morphological characters and substantiated their adaptation in high altitude as described earlier (van Dijk et al. Citation2004). The integrated approaches were rigorously adopted to delimitate the microhylid frogs and other closely related species throughout South and Southeast Asia and recognizing their phylogeny, origin, diversification, and colonization (Garg and Biju Citation2019; Gorin et al. Citation2020). In the recent past, combination with mitochondrial DNA (12S rRNA + 16S rRNA) and nuclear DNA (BDNF gene) has been extensively used to estimate the time-calibrated phylogeny of Microhyla frogs and discuss their taxonomy, biogeography, and evolution (Gorin et al. Citation2020). However, the mtCOI sequence information of microhylid frogs is still lacking in northeast India.
We generated both COI and 16S rRNA genetic information to elucidate the population genetic diversity of M. berdmorei among the different populations. The study provides the first DNA barcode data of the studied species from northeast India. The generated COI (MT755953) and 16S rRNA (MW578946) sequences depicted 96.43 and 99.64% similarity in nucleotide BLAST search with the published database sequences of M. berdmorei (MG935596 and MN534602) collected from Myanmar and Bangladesh respectively (Mulcahy et al. Citation2018; Gorin et al. Citation2020). The COI barcode and 16S rRNA sequences of M. berdmorei generated from distant localities were cohesively clustered in the BA phylogenies ().
For COI dataset, generated and database sequences revealed a total of 11 haplotypes with 92 mutational steps, and haplotype diversity (Hd) = 0.97 (). Two COI sequences generated from Myanmar (MG935594 and MG935597) and two sequences generated from Malaysia (MG386471 and MG386471) were represented by a single haplotype. For 16S rRNA dataset, generated and database sequences depicted a total of 19 haplotypes with 38 mutational steps, and haplotype diversity (Hd) = 0.93 (). The 16S rRNA sequence of the studied sample and six sequences (MH807388, MH807387, MH807386, MH807385, MH807384, MN534602) collected from northeast India and Bandarban, Bangladesh formed a single haplotype. Further, four sequences generated from Myanmar (MG935891, MG935890, MG935889, MG935888), two sequences generated from Thailand (MN534604, KR827906), two sequences generated from Thailand and Cambodia (KR827908, KU840572) and three sequences from Malaysia (MN534600, AB530638, AB598338) were represented by a single haplotype. The remaining 16S rRNA sequences were formed distinct haplotypes with sufficient mutational steps.
The studied sample collected from northeast India revealed genetically close with the Burmese collection with 3.8% genetic distance in COI gene. The northeast Indian sample also depicted high genetic distance (7.1–7.8% from Thailand and Laos samples and 11.2–11.8% from Malaysia samples in COI gene. Further, in 16S rRNA gene analysis, the studied sample collected from Mizoram state showed less genetic distances (0–0.9%) compared with the samples collected from different states (Assam and Tripura) in northeast India, Myanmar, and Bangladesh. The studied sample also depicted 1.7–4.6% genetic distance from other samples collected from different localities in Southeast Asian countries (Thailand, Vietnam, Laos, Cambodia, and Malaysia). A recent study reported a number of deep lineages (with >1.5% uncorrected genetic p-distances in 16S rRNA) within the different population of M. berdmorei collected from Southeast Asian countries and indicated potentially constituting cryptic species diversity (Gorin et al. Citation2020). Hence, considering the high intra-species genetic distances 3.8–11.8% in COI gene and 0–4.6% in 16S rRNA gene, this study corroborates the previous hypothesis and assumed the existence of possible cryptic diversity and species complex of M. berdmorei within its range distribution. However, this study hinted an additional sampling from board geographical regions and their genetic information would be useful to estimate the population genetics of this species precisely.
A recent study argued the confusing status of M. berdmorei types, and depicted the type locality in Bago, Myanmar (Garg et al. Citation2019). Besides, the species Microhyla fowleri was first described from Chieng Mai, Thailand, and later on synonymized and resurrected from M. berdmorei. Subsequently, the integrated taxonomic approaches neither elucidate the distinctiveness of M. fowleri, nor synonymized with M. berdmorei (Poyarkov et al. Citation2014; Garg et al. Citation2019). The most recent study illuminates the evolutionary position of M. fowleri under M. berdmorei group and noted as a complex of cryptic species (Gorin et al. Citation2020). Hence, the present study suggests the genetic information of both M. berdmorei and M. fowleri from their typical population which would be useful to discriminate the genetic distinctiveness and actual diversity in South and Southeast Asian countries.
Author's contribution
Conceptualization: SK, HTL; Sampling: LB, HD, LM; Data curation: SK, KT; Formal analysis: SK, KT, VK; Funding acquisition: KC, VK, HTL; Investigation: SK, HTL; Methodology: SK, LB; Project administration: KC, VK, HTL; Resources: KC; Software: SK, VK; Supervision: VK, KC, HTL; Validation: SK, VK; Visualization: SK, HTL; Writing – original draft: SK, HTL, KT; Writing – review and editing: SK, VK, HTL.
Acknowledgments
We are thankful to the Director of Zoological Survey of India (ZSI), Ministry of Environment, Forest and Climate Change (MoEF&CC), Govt. of India for providing necessary facilities and support for the study. The first author (SK) acknowledges the fellowship grant received from the Council of Scientific and Industrial Research (CSIR) Senior Research Associateship (Scientists’ Pool Scheme) Pool No. 9072-A. Authors would like to express their sincere thanks to the Chief Wildlife Warden, Department of Environment, Forest and Climate Change, Govt. of Mizoram for the collection permission, Mathipi Vabeiryureilai for helping in specimen collection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank database at (https://www.ncbi.nlm.nih.gov) with the accession numbers (MT755953 and MW578946) which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Additional information
Funding
References
- Biju SD, Garg S, Gururaja KV, Yogesh S, Sandeep WA. 2014. DNA barcoding reveals unprecedented diversity in dancing frogs of India (Micrixalidae, Micrixalus): a taxonomic revision with description of 14 new species. Ceylon J Sci (Biol Sci). 43(1):37–123.
- Biju SD, Garg S, Kamei RG, Maheswaran G. 2019. A new Microhyla species (Anura: Microhylidae) from riparian evergreen forest in the eastern Himalayan state of Arunachal Pradesh, India. Zootaxa. 4674(1):100–116.
- Chambers EA, Hebert PDN. 2016. Assessing DNA barcodes for species identification in North American reptiles and amphibians in natural history collections. PLoS One. 11(4):e0154363.
- Clement M, Snell Q, Walker P, Posada D, Crandall K. 2000. TCS: a computer program to estimate gene genealogies. Mol Ecol. 9(10):1657–1659.
- Das A, Garg S, Hamidy A, Smith EN, Biju SD. 2019. A new species of Micryletta frog (Microhylidae) from Northeast India. PeerJ. 7:e7012.
- De Bruyn M, Stelbrink B, Morley RJ, Hall R, Carvalho GR, Cannon CH, Van den Bergh G, Meijaard E, Metcalfe I, Boitani L, et al. 2014. Borneo and Indochina are major evolutionary hotspots for Southeast Asian biodiversity. Syst Biol. 63(6):879–901.
- De Sá RO, Streicher JW, Sekonyela R, Forlani MC, Loader SP, Greenbaum E, Richards S, Haddad CF. 2012. Molecular phylogeny of microhylid frogs (Anura: Microhylidae) with emphasis on relationships among New World genera. BMC Evol Biol. 12(1):241.
- Feng Y, Blackburn DC, Liang D, Hillis DM, Wake DB, Cannatella DC, Zhang P. 2017. Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the Cretaceous-Paleogene boundary. Proc Natl Acad Sci USA. 114(29):E5864–5870.
- Frost DR, Grant T, Faivovich J, Bain RH, Haas A, Haddad CFB, De Sá RO, Channing A, Wilkinson M, Donnellan SC, et al. 2006. The amphibian tree of life. Bull Am Mus Nat Hist. 297:1–291.2.0.CO;2]
- Frost DR. 2020. Amphibian Species of the World: an Online Reference. Version 6.0. American Museum of Natural History, New York, USA. [accessed 2021 February 10]. http://research.amnh.org/herpetology/amphibia/index.html.
- Garg S, Biju SD. 2019. New microhylid frog genus from Peninsular India with Southeast Asian affinity suggests multiple Cenozoic biotic exchanges between India and Sci Rep. 9(1):1906.
- Garg S, Suyesh R, Das A, Jiang J, Wijayathilaka N, Amarasinghe AT, Alhadi F, Vineeth KK, Aravind NA, Senevirathne G, Meegaskumbura M, et al. 2019. Systematic revision of Microhyla (Microhylidae) frogs of South Asia: a molecular, morphological, and acoustic assessment. Vertebr Zool. 69:1–71.
- Gorin VA, Solovyeva EN, Hasan M, Okamiya H, Karunarathna DMSS, Pawangkhanant P, de Silva A, Juthong W, Milto KD, Nguyen LT, et al. 2020. A little frog leaps a long way: compounded colonizations of the Indian Subcontinent discovered in the tiny Oriental frog genus Microhyla (Amphibia: Microhylidae). PeerJ. 8:e9411.
- Grosjean S, Ohler A, Chuaynkern Y, Cruaud C, Hassanin A. 2015. Improving biodiversity assessment of anuran amphibians using DNA barcoding of tadpoles. Case studies from Southeast Asia. C R Biol. 338(5):351–361.
- Hasan M, Islam MM, Kuramoto M, Kurabayashi A, Sumida M. 2014. Description of two new species of Microhyla (Anura: Microhylidae) from Bangladesh. Zootaxa. 3755(5):401–408.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003a. Biological identifications through DNA barcodes. Proc R Soc Lond B. 270(1512):313–322.
- Hebert PDN, Ratnasingham S, deWaard JR. 2003b. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci B: Biol Sci. 270:596–599.
- IUCN 2021. The IUCN Red List of Threatened Species. Version 2020.3. [accessed 2021 February 10]. https://www.iucnredlist.org.
- Kamei RG, San Mauro D, Gower DJ, Van Bocxlaer I, Sherratt E, Thomas A, Babu S, Bossuyt F, Wilkinson M, Biju SD. 2012. Discovery of a new family of amphibians from northeast India with ancient links to Africa. Proc R Soc B. 279(1737):2396–2401.
- Köhler J, Jansen M, Rodríguez A, Kok PJR, Toledo LF, Emmrich M, Glaw F, Haddad CFB, Rödel M, Vences M. 2017. The use of bioacoustics in anuran taxonomy: theory. Terminology, methods and recommendations for best practice. Zootaxa. 4251:1–124.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lalremsanga HT. 2011. Studies on the ecology, breeding behavior and development of ranid and Microhylid anurans prevalent in Mizoram [northeast India. Unpublished PhD Thesis]. p. 282. North Eastern Hills University, Shillong, Meghalaya, India. p. http://hdl.handle.net/10603/5520.
- Leigh JW, Bryant D. 2015. popart: full-feature software for haplotype network construction. Methods Ecol Evol. 6(9):1110–1116.
- Letunic I, Bork P. 2007. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23(1):127–128.
- Matsui M, Hamidy A, Belabut DM, Ahmad N, Panha S, Sudin A, Khonsue W, Oh H-S, Yong H-S, Jiang J-P, et al. 2011. Systematic relationships of Oriental tiny frogs of the family Microhylidae (Amphibia, Anura) as revealed by mtDNA genealogy. Mol Phylogenet Evol. 61(1):167–176.
- Mulcahy DG, Lee JL, Miller AH, Chand M, Thura MK, Zug GR. 2018. Filling the BINs of life: Report of an amphibian and reptile survey of the Tanintharyi (Tenasserim) Region of Myanmar, with DNA barcode data. ZK. 757:85–152.
- Nylander JAA. 2004. Mr.Modeltest v2, Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.
- Palumbi SR. 1996. Nucleic acids II: the polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK, editors. Molecular systematics. Sinauer Associates, Inc. p. 205–247.
- Peloso PLV, Frost DR, Richards SJ, Rodrigues MT, Donnellan S, Matsui M, Raxworthy CJ, Biju SD, Lemmon EM, Lemmon AR, et al. 2016. The impact of anchored phylogenomics and taxon sampling on phylogenetic inference in narrow-mouthed frogs (Anura, Microhylidae). Cladistics. 32(2):113–140.
- Poyarkov NA, Gorin VA, Zaw T, Kretova VD, Gogoleva SI, Pawangkhanant P, Che J. 2019. On the road to Mandalay: contribution to the Microhyla Tschudi, 1838 (Amphibia: Anura: Microhylidae) fauna of Myanmar with description of two new species. Zool Res. 40(4):244–276.
- Poyarkov NA, Nguyen TV, Duong TV, Gorin VA, Yang JH. 2018a. A new limestone-dwelling species of Micryletta (Amphibia: Anura: Microhylidae) from northern Vietnam. PeerJ. 6:e5771.
- Poyarkov NA, Suwannapoom C, Pawangkhanant P, Aksornneam A, Duong TV, Korost DV, Che J. 2018b. A new genus and three new species of miniaturized microhylid frogs from Indochina (Amphibia: Anura: Microhylidae: Asterophryinae). Zool Res. 39:130–155.
- Poyarkov NA, Vassilieva AB, Orlov NL, Galoyan EA, Tran DTA, Le DTT, Kretova VD, Geissler P. 2014. Taxonomy and distribution of narrow-mouth frogs of the genus Microhyla Tschudi, 1838 (Anura: Microhylidae) from Vietnam with descriptions of five new species. Russ J Herpetol. 21:89–148.
- Rakotoarison A, Scherz MD, Glaw F, Köhler J, Andreone F, Franzen M, Glos J, Hawlitschek O, Jono T, Mori A, et al. 2017. Describing the smaller majority: integrative taxonomy reveals twenty-six new species of tiny microhylid frogs (genus Stumpffia) from Madagascar. Vertebr Zool. 67:271–398.
- Rassmann K. 1997. Evolutionary age of the Galapagos iguanas predates the age of the present Galapagos Islands. Mol Phylogenet Evol. 7(2):158–172.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Rozas J, Ferrer-Mata A, Sanchez-DelBarrio J, Guirao-Rico S, Librado P, Ramos-Onsins S, Sanchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34(12):3299–3302.
- Seshadri KS, Singal R, Priti H, Ravikanth G, Vidisha MK, Saurabh S, Pratik M, Gururaja KV. 2016. Microhyla laterite sp. nov., a new species of Microhyla Tschudi, 1838 (Amphibia: Anura: Microhylidae) from a laterite rock formation in South West India. PLoS One. 11(3):e0149727..
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882.
- Tordoff AW, Bezuijen MR, Duckworth JW, Fellowes JR, Koenig K, Pollard EHB, Royo AG. 2011. Ecosystem Profile: Indo-Burma Biodiversity Hotspot, 2011 Update. Critical Ecosystem Partnership Fund. Washington, DC. p. 381.
- Tu N, Yang M-H, Liang D, Zhang P. 2018. A large-scale phylogeny of Microhylidae inferred from a combined dataset of 121 genes and 427 taxa. Mol Phylogenet Evol. 126:85–91.
- Van Bocxlaer I, Roelants K, Biju SD, Nagaraju J, Bossuyt F. 2006. Late Cretaceous vicariance in Gondwanan amphibians. PLoS One. 1(1):e74.
- van Dijk PP, Inger R, Iskandar D, Datong Y, Ohler A, Shunqing L, Dutta S, Bordoloi S, Sengupta S, Asmat GSM. 2004. Microhyla berdmorei (errata version published in 2018). The IUCN Red List of Threatened Species 2004: e.T57876A136565297.
- Vences M, Thomas M, Bonett RM, Vieites DR. 2005. Deciphering amphibian diversity through DNA barcoding: chances and challenges. Phil Trans R Soc B. 360(1462):1859–1868.
- Vieites DR, Wollenberg KC, Andreone F, Köhler J, Glaw F, Vences M. 2009. Vast underestimation of Madagascar’s biodiversity evidenced by an integrative amphibian inventory. Proc Natl Acad Sci USA. 106(20):8267–8272.
- Vineeth KK, Radhakrishna UK, Godwin RD, Anwesha S, Rajashekhar KP, Aravind NA. 2018. A new species of Microhyla Tschudi, 1838 (Anura: Microhylidae) from West Coast of India: an integrative taxonomic approach. Zootaxa. 4420(2):151–179.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding of Australia’s fish species. Phil Trans R Soc B. 360(1462):1847–1857.
