Abstract
The complete mitochondrial DNA (mtDNA) for Pareas stanleyi was determined in this study. The length of complete mtDNA was 18,932 bp, including 13 protein-coding genes (PCGs) (COI-III, ND1-6, ND4L, ATP6, ATP8 and CYTB), 23 tRNA genes, 2 rRNA genes, a L-chain replication-initiating non-coding region (NCR) and 2 control regions. The overall base composition of the sequence is 24.76% of T, 29.20% of C, 30.87% of A, and 15.16% of G, with a total A + T content of 55.63%. The phylogenetic tree showed that P. stanleyi had a close relationship with the other two species (P. boulengeri and P. formosensis) from the genus Pareas.
Pareas is a genus of Asian snakes in the family Pareidae, which contains 20 species (Bhosale et al. Citation2020). Pareas stanleyi is an endemic species, which is distributed in Fujian, Guizhou, Hunan, and Sichuan Provinces of China (Zhao Citation2006). In GenBank, only two complete mitochondrial DNA (mtDNA) are available for Pareas species, which are P. boulengeri (Huang et al. Citation2020) and P. formosensis (Liu et al. Citation2021). Here, we determined the complete circular mtDNA sequence of P. stanleyi via whole genomic sequencing, and found that it had a close relationship with the other species from the genus Pareas.
We collected the specimen (species voucher: LSU2020MLFJDT01) of P. stanleyi in Mount Wuyi National Park, from Nanping, Fujian Province, China (N27.71681°, E117.65694°) in July 2020. Presently, the specimen was stored in the Laboratory of Amphibian Diversity Investigation (ADI) at Lishui University (contact person: Li Ma, E-mail: [email protected]). Total genomic DNA was extracted from the muscle tissue of P. stanleyi using EasyPure Genomic DNA Kit (TransGen Biotech Co, Beijing, China). The whole genomic sequences was ultrasonic fragmented, then sequenced by Illumina NovaSeq 6000 platform (Novogene Bioinformatics Technology Co. Ltd., Tianjin, China) for paired-150 bp. Raw sequence data (15.57G) were deposited in NCBI’s Sequence Read Archive (SRA accession: SRR13500293). The NOVO Plasty 3.7 was used to de novo assembled the clean data without sequencing adapters (Dierckxsens et al. Citation2017).
The mtDNA of P. stanleyi (GenBank accession no. MW531673) is a closed-circular molecule of 18,932 bp in length, which including 13 PCGs (COX1-3, ND1-6, ND4L, ATP6, ATP8, and CYTB), 23 tRNA genes, 2 rRNA genes, a L-chain replication-initiating non-coding region, and 2 control regions. The composition and arrangement of P. stanleyi mtDNA were approximately the same as P. boulengeri (Huang et al. Citation2020) and P. formosensis (Liu et al. Citation2021). The overall base composition of the sequence is 24.76% of T, 29.20% of C, 30.87% of A, and 15.16% of G, with a total A + T content of 55.63%. There are two duplicate genes of tRNALeu and tRNASer in P. stanleyi mtDNA. All genes were encoded on the heavy strand except ND6 and eight tRNA genes (tRNAPro, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, and tRNAGlu), which were encoded on the light strand. Among the 13 PCGs, the ND5 was the longest (1788 bp), while the ATP8 was the shortest (162 bp). Codon usage analysis of P. stanleyi showed that four kinds of start codons (ATA, ATG, ATT, and GTG) and six kinds of stop codons (AGA, AGG, TAA, TAG, TA-, and T–) were used. The 23 tRNA genes varied in size from 56 to 73 bp. The 12S rRNA (920 bp) and 16S rRNA genes (1487 bp) are located between tRNAPhe and tRNAVal and between tRNAVal and tRNAIle, respectively.
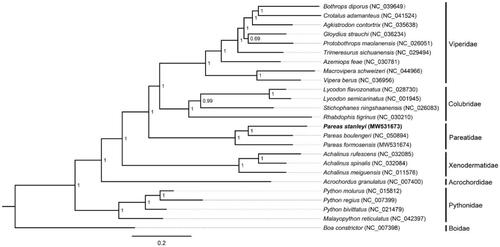
Phylogenetic analysis was inferred from available mtDNA of P. stanleyi and other 24 snakes based on 13 concatenated PCGs (11,382 bp) with Boa constrictor as the outgroup using Bayesian inference (BI) method in MrBayes v3.2.2. The optimal substitution model (GTR + I + G) was implemented by MrModelTest 2.3 (Nylander Citation2004). We analyzed four parallel runs of Markov Chain Monte Carlo (MCMC) for 1,000,000 generations, sampling every 1000 generations and discarded 1000 trees as burn-in. The phylogenetic tree showed that P. stanleyi was the sister species of P. boulengeri within the genus of Pareas (). The phylogenetic analysis result was consistent with the previous research. The genetic distance (p-distance) between P. stanleyi and P. formosensis, and between P. stanleyi and P. boulengeri was 20.16% and 18.44%, respectively, based on the 13 PCGs using MEGA 5.05 (Free Software Foundation, Boston, MA). The first determined mitogenome sequence of P. stanleyi will provide fundamental data for further exploring mDNA evolution in snakes.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitogenome data supporting this study are openly available in GenBank at (https://www.ncbi.nlm.nih.gov/nuccore/MW531673). Reference number (Accession no. MW531673). BioSample and SRA accession numbers are https://www.ncbi.nlm.nih.gov/biosample/SAMN17393257 and https://www.ncbi.nlm.nih.gov/sra/SRR13500293, respectively.
Additional information
Funding
References
- Bhosale H, Phansalkar P, Sawant M, Gowande G, Patel H, Mirza ZA. 2020. A new species of snail eating snakes of the genus Pareas Wagler, 1830 (Reptilia: Serpentes) from eastern Himalayas, India. Eur J Taxon. 729:54–73.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Huang RY, Peng LF, Yang DC, Yong Z, Huang S. 2020. Mitochondrial genome of the Boulenger's Slug eating snake Pareas boulengeri (Serpentes: Pareidae). Mitochondr DNA B. 5(3):3179–3180.
- Liu YL, Zhang YJ, Yang X, Yang XX, Lin ZH, Ma L. 2021. The complete mitochondrial genome of Taiwan slug-eating snake (Pareas formosensis) and phylogenetic analysis. Mitochondr DNA B. 6:970–971 (under review).
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.
- Zhao EM. 2006. Snakes of China. Hefei, China: Anhui Sciences and Technology Publishing House, p. 245–247 (in Chinese).