Abstract
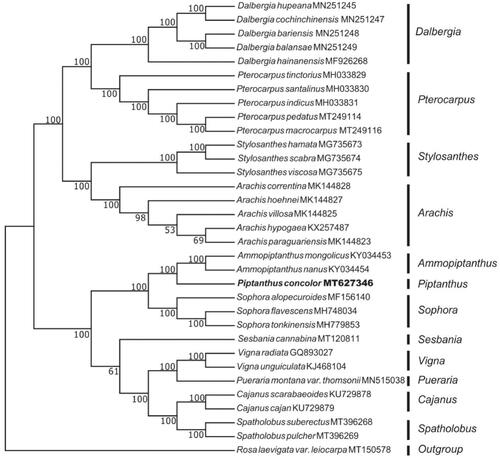
In this study, we used high-throughput sequencing technology to sequence and functionally annotate the chloroplast genome of Piptanthus concolor. The total length of P. concolor chloroplast genome is 152,115 bp with the typical quadripartite structure. It contains two inverted repeats (IRa and IRb) of 26,233 bp each, which are separated by a large single-copy (LSC) region of 82,024 bp and a small single-copy (SSC) region of 17,625 bp. The cp genome contained 111 genes, including 77 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis based on the whole chloroplast genome showed that P. concolor was closely related to Ammopiptanthus.
Piptanthus Sweet is a small genus in the family Leguminosae, three species and two forms of which have been found in China. Piptanthus concolor Craib is widely distributed in the region from the Himalaya to the Hengduan Mountains (Sun Citation2002). The seeds of P. concolor are used as Tibetan folk medicine (Liu and Jia Citation1991). Eleven isoflavonoids were isolated and identiffied from stem of P. concolor (kou et al. Citation2012). karyotype analysis of P. concolor was performed with fluorescence in situ hybridization(FISH) (Luo et al. Citation2017). However, there are rare reports about the chloroplast (cp) genome information of Piptanthus. In this study, we determined the complete chloroplast genome of P. concolor based on high throughput sequencing technology, which will provide bioinformatics data for the phylogeny of Piptanthus genus and further research on Leguminosae.
The fresh leaves of P. concolor were collected from Dali, Yunnan province, China (100°6’43.1′’E, 25°50’46.07′’N). The voucher specimen (NO: ZDQ17066) was deposited at the Herbarium of Medicinal Plants and Crude Drugs of the College of Pharmacy, Dali University. Total genomic DNA was extracted from the fresh leaves using a modified CTAB method (Doyle Citation1987; Yang et al. Citation2014). Genome sequencing was performed using Illumina Hiseq 2500 (Novogene, Tianjing, China) platform with pair-end (2 × 300bp) library. The raw data have filtered using Trimmomatic version 0.32 with default settings (Bolger et al. Citation2014). Then the filtered reads were assembled into circular contigs using GetOrganelle (Jin et al. Citation2020), with the complete chloroplast genome of Ammopiptanthus mongolicus as the reference (GenBank accession no. KY034453). Finally, the chloroplast genomes were annotated using Geneious R11.0.2 (Kearse et al. Citation2012), and corrected manually for start and stop codons. The complete chloroplast genome sequence of P. concolor was submitted to GenBank with the accession number of MT627346.
The complete chloroplast genome of P. concolor is 152,115 bp in length with the typical quadripartite structure, which consists of a large single copy-region (LSC, 82,024 bp), a small single copy-region (SSC, 17,625 bp), and a pair of inverted repeat regions (IRa and IRb, 26,233 bp). The full-sequence GC content of the chloroplast genome was 36.6%. 111 chloroplast genes were successfully annotated, including 4 rRNA genes, 30 tRNA genes and 77 protein-coding genes. 12 genes contain introns, among which 10 genes have only one intron and 2 genes (ycf3 and clpP) have two introns.
In order to determine the phylogenetic position of P. concolor, we selected 32 chloroplast genome sequences from NCBI. All sequences were aligned using program MAFFT v.7.149 (Katoh and Standley Citation2013). and a neighbour-joining (NJ) phylogenetic tree was constructed by MEGA v.7.0.26 (Kumar et al. Citation2016) with 1000 bootstrap replicates. The phylogenetic analysis showed that P. concolor was closely related to Ammopiptanthus ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence date that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT627346.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CM, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organlle genomes. Genome Biol. 21: 241.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kou ZJ, Chang RJ, Qin JJ, Yan SK, Jin HZ, Zhang WD. 2012. Isoflavonoids from Piptanthus concolor Harrow. NatProd Res Dev. 24(5):610–613.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu ZM, Jia ZJ. 1991. Studies on the chemical constituents of Piptanthus concolor Harrow. Chem J Chin Univ. 12:774–776.
- Luo XM, Liu JC, Zhao AJ, Chen XH, Wan WL, Chen L. 2017. Karyotype analysis of Piptanthus concolor based on FISH with a oligonucleotides for rDNA 5S. Sci Hortic. 226(12):361–365.
- Sun H. 2002. Tethys retreat and Himalayas-Hengduanshan mountains uplift and their significance on the origin and development of the Sino-Himalayan elements and alpine flora. Acta Bot Yunnan. 3:273–288.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.