Abstract
Gynaikothrips ficorum (Marchal Citation1908) is a major pest of bonsai ficus and poses a considerable economic threat to gardening industry. The mitochondrial genome of G. ficorum was sequenced and annotated in this study. Its whole mitogenome was 15,313 bp in length, including 37 typical genes in animal mitogenomes. ATN was used as start codon in most of the PCGs except for nad4l, which used TTG. All PCGs used TAA as termination codon except atp8 and atp6 which were ended with an incomplete T and TAG, respectively. A phylogenetic tree based on complete mitochondrial genomes of 17 species (15 Thysanoptera species and two outgroups) showed that the monophyly of Phlaeothripidae was supported and G. ficorum and G. uzeli formed a sister group.
A major pest of bonsai ficus, Gynaikothrips ficorum (Marchal Citation1908) preferentially feeds on tender young leaves and induces leaf-fold galls on Ficus microcarpa (Okajima Citation2006). The infested trees will not be killed, but the ornamental value and quality of the plants are reduced markedly (Han Citation1997). In this study, the samples of G. ficorum were collected from F. microcarpa located beside the road at Yibin City in Sichuan Province, China (104°60′04″E, 28°75′14″N) in 2015. Fresh specimens were preserved in 95% ethanol for slide mounting and DNA extraction. Voucher specimens (JM2015051) were identified by morphology, and deposited in Qinba Biological Museum, Shaanxi University of Technology, Hanzhong, China (QBM). The genomic DNA extraction of G. ficorum is used by the DNeasy kit (Qiagen, Hilden, Germany). The paired end libraries were constructed and sequenced (2 × 150bp) using the Illumina HiSeq 4000 platform by Nextomics Bioscience Company (Wuhan, China). The complete mitogenome for this species was assembled and annotated by MITOZ (Meng et al. Citation2019).
The complete mitogenome of G. ficorum is a typical circular double-stranded DNA molecule of 15,313 bp in length (GenBank accession no. MT892761). It consists of 13 protein-coding genes (PCGs), 22 transfer RNA genes in which trnM and trnY are duplicated, two ribosomal RNA genes and one control region (d-loop). Twenty-four genes (4 PCGs, 18 tRNA and 2 rRNA genes) are transcribed on the majority strand (J-strand), with the remaining genes being located on the minority strand (N-strand). The overall nucleotide composition of the G. ficorum mitogenome is 42.8% T, 8.6% C, 40.3% A and 8.3% G, with a strong bias toward A + T (83%), AT- and GC- skew of the whole mitogenome of G. ficorum is −0.029 and −0.017, respectively. Most PCGs start with ATN codon, expect nad4l, which starts with TTG codon. Most PCGs were ended with TAA codon expect atp8 and atp6 which are ended with an incomplete T and TAG, respectively. A single putative control region, rather than two or more in some other thrips species (Yan et al. Citation2012; Shao and Baker Citation2003; Yan et al. Citation2014; Dickey et al. Citation2015; Chakraborty et al. Citation2018; Kumar et al. Citation2019), is found in the mt genome of G. ficorum. The unique control region is 251 bp in length and located between nad5 and trnF. The secondary structure of tRNA were predicted by MITOS (Bernt et al. Citation2013) and found that except trnS1 loses a dihydrouridine (DHU) arm, all others form a typical clover-leaf structure.
Comparing the arranged mitochondrial genes of G. ficorum with inferred insect ancestral arrangement, we found severe rearrangements in the G. ficorum mitochondrial genome. Most genes arranged irregularly but two gene clusters (atp8-atp6-cox3 and nad4-nad4L) conserved. The mt genes of this target species arranged in cox1-trnQ-cox2-atp8-atp6-cox3-nad3-trnN-trnS1-trnE-nad5-trnF-trnH -trnS-trnC-nad4-nad4L-trnT-nad6-cytb-trnR-trnA-trnY2-rrnS-trnD-trnG-trnL1-trnW-trnM1-trnY1-trnM2-trnV-trnK-trnI-nad2-trnL2-rrnL-trnP-nad1.
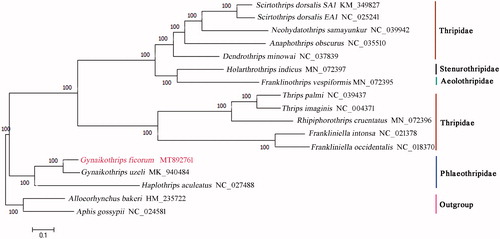
A phylogenetic tree was constructed based on the complete mitochondrial genomes of 15 Thysanoptera species together with two species Aphis gossypii and Alloeorhynchus bakeri as outgroups using the maximum-likelihood (ML) method with 1000 bootstrap replicates by MEGA7.0 (Sudhir et al. Citation2016) (). The phylogenetic relationships indicated by the tree supported completely their taxonomic relationships based on morphological characteristic (ThripsWiki Citation2020), in which G. ficorum and G. uzeli were clustered into a clade, and they formed a sister clade to H. aculeatus. The result showed the three species belonging to two genera have close relationships, and it supported the monophyly of Phlaeothripidae.
Disclosure statement
The authors declare that they have no conflict of interest.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT892761. The associated SRA, BioProject and Bio-Sample numbers are as follows: SRR14205964, PRJNA721109, and SAMN18700104.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chakraborty R, Tyagi K, Kundu S, Kundu S, Rahama I, Singha D, Chandra K, Patnaik S, Kumar V. 2018. The complete mitochondrial genome of Melon thrips, Thrips palmi (Thripinae): comparative analysis. PLoS One. 13(10):e0199404.
- Dickey AM, Kumar V, Morgan JK, Jara-Cavieres A, Shatters RG, Jr, McKenzie CL, Osborne LS. 2015. A novel mitochondrial genome architecture in thrips (Insecta: Thysanoptera): extreme size asymmetry among chromosomes and possible recent control region duplication. BMC Genomics. 16(1):439.
- Han YF. 1997. Economic Insect Fauna of China. 55 Thysanoptera. Beijing: Editorial Committee of Fauna Sinica, Science Press; 514pp.
- Kumar V, Tyagi K, Kundu S, Chakraborty R, Singha D, Chandra K. 2019. The first complete mitochondrial genome of marigold pest thrips, Neohydatothrips samayunkur (Sericothripinae) and comparative analysis. Sci Rep. 9(1):191.
- Marchal P. 1908. Sur une nouvelle espece de Thrips nuisible aux Ficus on Algerie. Bulletin de la Societe Entomologique de France. 77:251–253.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Okajima S. 2006. The insects of Japan. Vol. 2. The suborder Tubulifera (Thysanoptera). Fukuoka: Touka Shobo Co Ltd.; 720pp.
- Shao R, Barker SC. 2003. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol Biol Evol. 20(3):362–370.
- Sudhir K, Glen S, Koichiro T. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 7:1870.
- ThripsWiki. 2020. ThripsWiki—providing information on the World’s thrips. Available from: http://thrips.info/wiki/Main_Page [accessed 2020 July 20].
- Yan DK, Tang Y, Hu M, Liu FQ, Zhang DF, Fan JQ. 2014. The mitochondrial genome of Frankliniella intonsa: insights into the evolution of mitochondrial genomes at lower taxonomic levels in Thysanoptera. Genomics. 104(4):306–312.
- Yan DK, Tang YX, Xue XF, Wang MH, Liu FQ, Fan JQ. 2012. The complete mitochondrial genome sequence of the western flower thrips Frankliniella occidentalis (Thysanoptera: Thripidae) contains triplicate putative control regions. Gene. 506(1):117–124.