Abstract
Phalaenopsis wilsonii Rolfe is a vulnerable wild moth orchid species with important horticultural value. The complete chloroplast genome sequence of P. wilsonii was generated by de novo assembly using whole genome next-generation sequencing to provide genomic data for further conservation genetics, phylogeny and molecular breeding in Phalaenopsis. The complete plastome of P. wilsonii is 145,096 bp in length, containing two inverted repeats (IR) regions (24,787 bp), a large single-copy (LSC) region (84,688 bp), and a small single-copy (SSC) region (10,834 bp). The chloroplast genome encoded 119 unique genes, including 73 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The overall GC content of the whole genome is 36.9%. Phylogenetic analysis indicated P. wilsonii was closely related to P. lowii.
Phalaenopsis (Vandeae, Orchidaceae) is one of the most important and popular ornamental flowers in the world because of its beautiful appearance and high ornamental value. Based on morphology and DNA evidence, the Phalaenopsis genus is divided into four subgenera, namely Parishianae, Phalaenopsis, Hygrochilus, and Ornithochilus (Christenson Citation2001; Kocyan and Schuiteman Citation2014; Li et al. Citation2016). Phalaenopsis wilsonii Rolfe is belonging to subgenera Parishianae, which grow on trees or damp rocks under forests, mainly distributed among southwest China and northern India. Phalaenopsis is principally inhabiting the tropical regions, while P. wilsonii is also distributed in temperate zone. With purple pink flowers and abundant aerial roots, P. wilsonii is an important breeding parent of Phalaenopsis (Liu Citation2008). Phalaenopsis wilsonii has been categorized as vulnerable in China Species Red List (Wang and Xie Citation2004). Here, we report the complete chloroplast genome of P. wilsonii, which will provide genetic and genomic information for further conservation genetics, phylogenetic studies and future breeding in Phalaenopsis.
Fresh leaves of P. wilsonii were collected from Shangri-La county in northwestern Yunan province of China (99°29′31.41″E, 27°48′7.88″N, 2241 m). Voucher specimen (SWFU20200711MFY) was deposited in the Herbarium of Southwest Forestry University, China. Total genomic DNA was extracted from its fresh leaves using the Axygen® AxyPrep Multisource Genomic Miniprep DNA kit (Corning, Corning, NY) according to the manufacturer’s instruction. A pair-end (PE) library was constructed and sequenced using the Illumina HiSeq 2500-PE150 platform (Illumina, San Diego, CA). The clean reads was obtained from filtered raw reads using NGS QC Toolkit_v2.3.3 with default parameters (Patel and Jain Citation2012). The plastome was de novo assembled by NOVOPlasty (Dierckxsens et al. Citation2017), and annotated by Geneious Prime (Kearse et al. Citation2012) with the complete chloroplast genome sequence of P. japonica (NC_046808) as the reference. The complete chloroplast genome of P. wilsonii was submitted to GenBank with accession number MW194929.
The complete plastome of P. wilsonii is 145,096 bp in length, containing a large single-copy (LSC) region of 84,688 bp, a small single-copy (SSC) region of 10,834 bp, and a pair of inverted repeats (IR) regions of 24,787 bp. The overall GC content of the whole genome is 36.9%. In total, 119 unique genes were annotated, including 73 protein-coding genes (PCGs), 8 ribosomal RNA genes (rRNAs), and 38 transfer RNA genes (tRNAs). A total of 69 SSRs were discovered by the online software MISA-web (Beier et al. Citation2017). Among them, the numbers of mono-, di-, tri-, tetra- and penta-nucleotides SSRs are 49, 7, 4, 7, and 2, respectively.
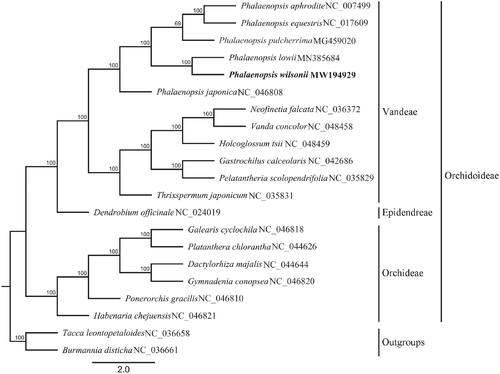
To confirm the phylogenetic position of P. wilsonii, other 18 published complete chloroplast genomes from Orchidaceae were aligned by using MAFFT v.7 (Katoh and Standley Citation2013). Tacca leontopetaloides and Burmannia disticha were used as outgroups. A maximum-likelihood tree () was constructed with RAxML v8.2.11 (Stamatakis Citation2014) in which the GTR + G DNA substitution model was selected and all branch nodes were calculated under 1,000 bootstrap replicates. The phylogenetic tree showed that P. wilsonii was closely related to P. lowii. The result also appears in the ML tree constructed with ITS sequences (Tsai et al. Citation2003) and the combined plastid DNA (Tsai et al. Citation2010). The complete chloroplast genome sequence of P. wilsonii will provide useful information for further study on conservation genetics, phylogeny and molecular breeding in Phalaenopsis even in Orchidaceae.
Authors contributions
C. L. M conceived the study; D. Y. Y. collected the molecular materials; Z. F. F. drafted the manuscript; C. L. M. revised the manuscript. All authors provided comments and final approval.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/, reference number (MW194929) (SRR12929239), or obtain from the corresponding author.
Additional information
Funding
References
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585.
- Christenson EA. 2001. Phalaenopsis: a monograph. Portland, OR: Timber Press.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kocyan A, Schuiteman A. 2014. New combinations in Aeridinae (Orchidaceae). Phytotaxa. 161(1):61–85.
- Li MH, Gruss O, Liu ZJ. 2016. Nomenclature changes in Phalaenopsis subgen. Hygrochilus (Orchidaceae; Epidendroideae; Vandeae) based on DNA evidence. Phytotaxa. 275(1):55–61.
- Liu YT. 2008. Orchid: aesthetic, application, cultivation. Beijing (China): China Forestry Press.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tsai CC, Chiang YC, Huang SC, Chen CH, Chou CH. 2010. Molecular phylogeny of Phalaenopsis blume (orchidaceae) on the basis of plastid and nuclear DNA. Plant Syst Evol. 288(1-2):77–98.
- Tsai CC, Huang SC, Huang PL, Chou CH. 2003. Phylogeny of the genus Phalaenopsis (orchidaceae) with emphasis on the subgenus Phalaenopsis based on the sequences of the internal transcribed spacers 1 and 2 of rdna. Taxon. 52(6):287–294.
- Wang S, Xie Y. 2004. China species red list. Beijing (China): Higher Education Press.