Abstract
The chloroplast (cp) genome sequence of Rhizophora apiculata was characterized. The cp genome length was 164,343 bp in length, containing a typical structure of a large single copy (LSC) of 93,155 bp, a small single copy (SSC) of 19,376 bp, and two inverted repeats (IRs) of 25,906 bp, with a GC content of 34.9%. There were 131 genes were annotated in the cp genome, including 85 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. A phylogenetic analysis using cp genomes of mangroves and ecologically associated species resolved R. apiculata in Rhizophora with R. stylosa and R. x lamarckii. This complete chloroplast sequence offers a promising tool for further species identification and evolutionary studies of Rhizophora, as well as for mangroves.
Introduction
Mangroves are a diverse group of woody trees and shrubs that thrive in saline and daily inundation conditions in the inter-tidal zone of tropical and subtropical regions (Ball Citation1988; Duke Citation1992). Mangrove ecosystems are among the most carbon-rich forests in the tropics (Donato et al. Citation2011; Alongi Citation2014), not only providing the services for lots of animals, but also providing protections from erosion, storm surge, and potentially tsunami waves (Gedan et al. Citation2011). Mangroves are threatened by climate changed-induced drought, as well as, relative sea level rise (Lovelock Citation2020; Saintilan et al. Citation2020). Mangrove forests have been severely degraded over the past half century (Blasco et al. Citation2001; Duke et al. Citation2007; Donato et al. Citation2011), injecting new urgency into understanding the bio-resources of mangroves.
Rhizophora consists of five species and at least 6 hybrid taxa, which are largely separated into Old and New World groups (Schwarzbach Citation2014; Chen et al. Citation2015). The Old World taxa (R. apiculata, R. mucronata, R. stylosa, R. x annamalayana, R. x lamarckii) are considered the most important of all mangroves across the Pacific tropical region (Duke Citation2006, Citation2010). Therefore, providing the genome sequences of species R. apiculata will help to spur research on their interesting adaptations, and also could offer some needed information for its conservation. Chloroplast DNA (cpDNA) could provide useful and abundant information on genetic diversity and evolution based on our previous studies (Jiang et al. Citation2016; Lei et al. Citation2018), as well as in other mangrove species (Chen et al. Citation2019; Yang et al. Citation2019; Shi et al. Citation2020). In this study, we assembled and characterized the chloroplast genome of R. apiculata based on Illumina pair-end data, and built a phylogenetic tree using plastomes available in mangroves and ecologically associated species.
Total genomic DNA was extracted from frozen fresh leaves collected in Sanya Tielu Port mangrove reserve (Sanya, P. R. China, 18° 15′ N/109° 42′ E), following previously described methods (Jiang et al. Citation2016; Lei et al. Citation2018). The voucher (R. apiculata_Jiang_R9) is stored in Guangxi University, plant ecophysiology and evolution research group herbarium. A 350-bp paired-end library was then constructed and sequenced by Novogene (Beijing, PR China) using an Illumina HiSeqX-ten system (Illumina, San Diego, CA). After sequence filtering for read quality, about 1 Gb raw data were obtained, with a paired-end read length of 2 × 150 bp. A de novo assembly was performed using NOVOPlasty3.6 (Dierckxsens et al. Citation2016), using a seed sequence from the mangrove Avicennia marina (AB114520.1). The assembled sequence was then imported into Geneious R9 (Biomatters Ltd, Auckland, New Zealand), to check manually as described previously (Jiang et al. Citation2016; Hinsinger and Strijk Citation2017; Xu et al. Citation2017, Citation2018). The minimum coverage for the assembled cp genome was 48 ×, and annotations were then transferred from Rhizophora x lamarkii (NC_046517). The final annotations were confirmed and integrated from results of CPGAVAS2 (Shi et al. Citation2019) and IRscope (Amiryousefi et al. Citation2018).
The assembled cp genome of R. apiculata had a length of 164,343 bp (GenBank accession number MW387538). The cp genome exhibited a typical structure with a LSC of 93,155, a SSC 19,376 and two inverted repeats (IRa and IRb) of 25,906 bp, respectively. The overall GC content of the plastome was 34.9%, while the GC content in LSC, SSC, IRa, and IRb regions were 32.2%, 28.5%, 42.2%, respectively. In total, 131 genes were annotated, including 85 protein-coding genes, 38 tRNA genes, and 8 ribosomal RNA genes. Seventeen genes (atpF, clpP, petB, petD, rpl16, rps12, rpoC1, ycf3, trnG-UCC, trnK-UUU, trnL-UAA, trnV-UAC in LSC; ndhA locates in SSC; ndhB, rpl2, trnA-UGC, trnI-GAU in the IRs regions) contain 1–2 introns, respectively. While 17 genes were duplicated in the IR regions, including 6 protein-coding genes (rpl2, rpl23, ycf1, ycf2, ndhB, rps7), 7 tRNA genes (trnA-UGC, trnI-GAU, trnI-CAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC), and 4 rRNA genes (4.5S, 5S, 16S, 23S). Notably, sequence of the translation initiation factor gene (infA) was found located within rps8, with the existence of several internal termination codons indicating a functional loss of infA in the cp genome of R. apiculata.
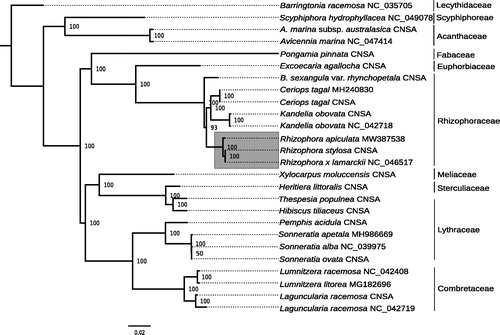
Twenty-four plastomes of mangroves and ecologically associated species were retrieved from GenBank (accessed 2020/11/17), and CNSA (https://db.cngb.org/cnsa/) according to the reference (Shi et al. Citation2020), plus Barringtonia racemosa as an out-group (). All the cp genome sequences were aligned with MAFFT v7.307 (Katoh and Standley Citation2013), and a maximum likelihood (ML) tree was built using PhyML v3.3 (Guindon et al. Citation2009) with a GTR + I + G model and support estimated with 1000 bootstraps replicates. All but one node were highly supported (bootstrap support ≥93), and R. apiculata clustered with the other two species of Rhizophora. The results indicated that providing the plastome of R. apiculata will be useful bioresource that will help to assess species identification and further genetic studies in mangroves.
Disclosure statement
The authors report no conflicts of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW387538. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA713533, SRR13933380, and SAMN18253766, respectively. Data were also available in the database: CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with accession number CNP0001525 (https://db.cngb.org/search/project/CNP0001525/).
Additional information
Funding
References
- Alongi DM. 2014. Carbon cycling and storage in mangrove forests. Annu Rev Mar Sci. 6(1):195–219.
- Amiryousefi A, Hyvonen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34(17):3030–3031.
- Ball MC. 1988. Ecophysiology of mangroves. Trees. 2(3):129–142.
- Blasco F, Aizpuru M, Gers C. 2001. Depletion of the mangroves of Continental Asia. Wetl Ecol Manag. 9(3):255–266.
- Chen Y, Hou Y, Guo Z, Wang W, Zhong C, Zhou R, Shi S. 2015. Applications of multiple nuclear genes to the molecular phylogeny, population genetics and hybrid identification in the mangrove genus rhizophora. PloS One. 10(12):e0145058.
- Chen Y, Yang Y, Li J, Jin Y, Liu Q, Zhang Y. 2019. The complete chloroplast genome sequence of a medicinal mangrove tree Ceriops tagal and its phylogenetic analysis. Mitochondrial DNA Part B. 4(1):267–268.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Donato DC, Kauffman JB, Murdiyarso D, Kurnianto S, Stidham M, Kanninen M. 2011. Mangroves among the most carbon-rich forests in the tropics. Nature Geosci. 4(5):293–297.
- Duke N. 2006. Indo-West Pacific stilt mangrove. Rhizophora apiculata, R. mucronata, R. stylosa, R. X annamalai, R. X lamarckii. In: Traditional trees of pacific islands: their culture, environment, and use. p. 641–660. Permanent Agriculture Resources, Hōlualoa, Hawaii, USA.
- Duke NC. 1992. Mangrove floristics and biogeography. In: AI R, DM A, editors. Tropical mangrove ecosystems. Washington (DC): American Geophysiocal Union; p. 63–100.
- Duke NC. 2010. Overlap of eastern and western mangroves in the South-western Pacific: hybridization of all three Rhizophora (Rhizophoraceae) combinations in New Caledonia. Blum J Plant Tax Plant Geog. 55(2):171–188.
- Duke NC, Meynecke J-O, Dittmann S, Ellison AM, Anger K, Berger U, Cannicci S, Diele K, Ewel KC, Field CD, et al. 2007. A world without mangroves? Science. 317(5834):41b–42b.
- Gedan KB, Kirwan ML, Wolanski E, Barbier EB, Silliman BR. 2011. The present and future role of coastal wetland vegetation in protecting shorelines: answering recent challenges to the paradigm. Clim Change. 106(1):7–29.
- Guindon S, Delsuc F, Dufayard J-F, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. In: Posada D, editor. Bioinformatics for DNA sequence analysis. Totowa (NJ): Humana Press; p. 113–137.
- Hinsinger DD, Strijk JS. 2017. Toward phylogenomics of Lauraceae: the complete chloroplast genome sequence of Litsea glutinosa (Lauraceae), an invasive tree species on Indian and Pacific Ocean islands. Plant Gene. 9:71–79.
- Jiang GF, Hinsinger DD, Strijk JS. 2016. Comparison of intraspecific, interspecific and intergeneric chloroplast diversity in Cycads. Sci Rep. 6(1):31473–31482.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Lei JY, Hinsinger DD, Jiang GF. 2018. Characterization of the complete chloroplast genome of endangered Cycads Zamia fischeri Miq. ex Lem. Mitochondrial DNA B Resour. 3(2):1059–1061.
- Lovelock CE. 2020. Blue carbon from the past forecasts the future. Science. 368(6495):1050–1052.
- Saintilan N, Khan NS, Ashe E, Kelleway JJ, Rogers K, Woodroffe CD, Horton BP. 2020. Thresholds of mangrove survival under rapid sea level rise. Science. 368(6495):1118–1121.
- Schwarzbach AE. 2014. Rhizophoraceae. In: Kubitzki K ed. The families and genera of vascular plants. Flowering plants. Eudicots. Malpighiales. Berlin, Heidelberg, Germany: Springer-Verlag; p. 283–295.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Shi C, Han K, Li L, Seim I, Lee SM-Y, Xu X, Yang H, Fan G, Liu X. 2020. Complete chloroplast genomes of 14 mangroves: phylogenetic and comparative genomic analyses. BioMed Res Inter. 2020:1–13.
- Xu LM, Hinsinger DD, Jiang G-F. 2017. The complete mitochondrial genome of the Agrocybe aegerita, an edible mushroom. Mitochondrial DNA B Resour. 2(2):791–792.
- Xu LM, Hinsinger DD, Jiang G-F. 2018. The complete mitochondrial genome of the Basidiomycete fungus Pleurotus cornucopiae (Paulet) Rolland. Mitochondrial DNA B Resour. 3(1):73–75.
- Yang Y, Zhang Y, Chen Y, Gul J, Zhang J, Liu Q, Chen Q. 2019. Complete chloroplast genome sequence of the mangrove species Kandelia obovata and comparative analyses with related species. PeerJ. 7:e7713.