Abstract
The number of species in the genus Pseudo-nitzschia has increased to 56, including 26 species known to produce domoic acid (DA), which is harmful to marine animals and human health. The lack of genomic sequences of Pseudo-nitzschia species has been a limiting factor in the studies of genetic and evolutionary relationships of Pseudo-nitzschia species. Here, the complete mitochondrial genome sequence of Pseudo-nitzschia micropora was determined for the first time, which was 38,792 bp in length with the overall AT content being 69.98%. The mitochondrial genome encoded 62 genes, including 36 protein-coding genes (PCGs, including orf157), 24 transfer RNA (tRNA) genes and two ribosomal RNA (rRNA) genes. Phylogenetic tree analysis suggests that the P. micropora had a closer relationship with P. cuspidate than that with P. multiseries. The availability of the complete mitochondrial genome of P. micropora would be useful for researching the evolutionary relationships of Pseudo-nitzschia species.
Many species of the genus Pseudo-nitzschia are cosmopolitan and can form blooms in coastal regions, some of which can produce domoic acid (DA) that cause amnesic shellfsh poisoning (ASP), which is harmful to seabirds, marine mammals and human health (Scholin et al. Citation2000; Dong et al. Citation2020). As a genus in the order Bacillariales (Bacillariophyceae, Bacillariophyta), the number of described Pseudo-nitzschia species has increased to 56 (Dong et al. Citation2020), including 26 known to produce DA (Dong et al. Citation2020). Nevertheless, the genomic sequences of these species are limited. Such genomic sequences are critical for analyzing genetic and evolutionary relationships of species in the genus Pseudo-nitzschia. Pseudo-nitzschia micropora Priisholm et al. Citation2002 is a nontoxic species but has been reported to be high abundance during an algal bloom in Cuyutlan Lagoon (Colima, Mexico) (Quijano-Scheggia et al. Citation2011). It has been identified in many countries including Thailand (Priisholm et al. Citation2002), Vietnam (Larsen and Nguyen Citation2004), Malaysia (Lim et al. Citation2012), Korea (Park et al. Citation2009) and China (Xu and Li Citation2015). In this study, we constructed the complete mitochondrial genome of P. micropora. The strain CNS00133 analyzed in this study was isolated from water sample using single cell capillary method, during an expedition to the Jiaozhou Bay (36°09.472′N, 120°15.055′ E) in August 2019 on the research vehicle ‘Innovation’, and its specimen was deposited in the collection of marine algae in KLMEES of IOCAS (Nansheng Chen, [email protected]) with the voucher number CNS00133. Briefly, after 2 weeks in culture, the algal cells were harvested by centrifugation (12,000 g, 5 min) and stored in liquid nitrogen for the next DNA extraction step.
Total genomic DNA of P. micropora was extracted using a TIANGEN DNAsecure Plant Kit (Tiangen Biotech, China) according to the manufacturer’s instructions. The genomic DNA library was sequenced using Illumina NovaSeq 6000 platform (Illumina, USA). Illumina sequencing results (about 5 Gb) of P. micropora were assembled into the mitochondrial genome using GetOrganelle v 1.7.2 (Jin et al. Citation2020), with SPAdes v 3.13.2 as the assembler (Bankevich et al. Citation2012). The completed mitogenome of P. micropora was circular in shape and 38,792 bp in length (Genbank accession number MW423602), which was slightly longer than that of P. cuspidate (37,203 bp), but much shorter than that of P. multiseries (46,283 bp) (Yuan et al. Citation2016). The overall AT content of the mitogenome of P. micropora was 69.98%, which was similar to that of P. cuspidate (69.83%) and P. multiseries (68.95%) (Yuan et al. Citation2016). The P. micropora mitogenome encoded 62 genes, including 36 protein-coding genes (PCGs, including orf157), 24 transfer RNA (tRNA) genes and two ribosomal RNA (rRNA) genes. The length of cox1 (1,512 bp) of P. micropora was identical to that of P. cuspidate. No introns were found in any genes of the P. micropora mitogenome. However, the tatA gene of P. micropora (129 bp) was much shorter than that of P. cuspidata (246 bp) and P. multiseries (234 bp). These three mitogenomes of the genus Pseudo-nitzschia shared 34 PCGs, while rps7 is absent from the mitochondrial genome of P. multiseries. There was no difference among the number and category of the tRNAs and rRNAs between three mitochondrial genomes, respectively.
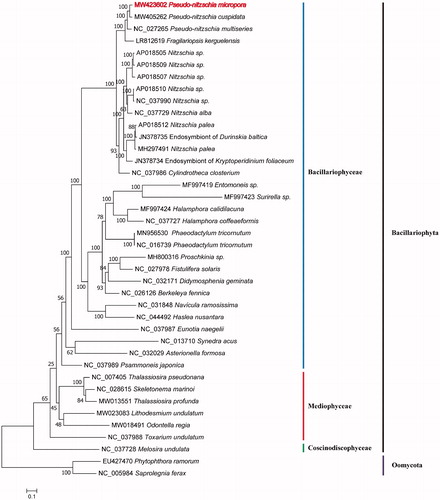
Maximum likelihood (ML) phylogenetic tree was constructed using tandem amino acid sequences of 31 common protein-coding genes (atp6, 8, 9; cob; cox1, 2, 3; nad1-7, 4 L, 9, 11; rpl2, 5, 6, 14, 16; rps3, 4, 8, 10, 11, 13, 14, 19; and tatC) shared by 40 species () using IQtree v1.6.12 (Trifinopoulos et al. Citation2016) with 1000 bootstrap alignments. Two Oomycota species Phytophthora Ramorum (EU427470) and Saprolegnia Ferax (NC_005984) were used as out-group taxa. The 38 Bacillariophyta species clustered well into three clades corresponding to three classes Coscinodiscophyceae, Mediophyceae and Bacillariophyceae. P. micropora clustered well with P. cuspidate and P. multiseries with strong support (). P. micropora had a closer relationship with P. cuspidate than P. multiseries.
Figure 1. Maximum likelihood (ML) tree based on 31 PCGs of 38 Bacillariophyta species plus two Oomycota species as outgroups. GenBank accession numbers are given before the species name.
The construction of the mitochondrial genome of P. micropora was not only valuable for studies on genetic diversity of this species, but also could contribute to further understanding of the evolutionary relationships of Pseudo-nitzschia species.
Acknowledgements
This cruise was conducted onboard R/V ‘Xiang Yang Hong 18’ by The First Institute of Oceanography, Ministry of Natural Rresources, China. Data acquisition and sample collections from the East China Sea were supported by National Natural Science Foundation of China (NSFC) Open Research Cruise (Cruise No. NORC2019-2), funded by Shiptime Sharing Project of NSFC.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW423602, under the accession no. MW423602.
The associated BioProject, SRA and Bio-Sample numbers are PRJNA687399 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA687399), SRR13319799 (https://www.ncbi.nlm.nih.gov/sra/SRR13319799) and SAMN17173859 (https://www.ncbi.nlm.nih.gov/biosample/ SAMN17173859), respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Dong HC, Lundholm N, Teng ST, Li A, Wang C, Hu Y, Li Y. 2020. Occurrence of Pseudo-nitzschia species and associated domoic acid production along the Guangdong coast, South China Sea. Harmful Algae. 98(101899):101899.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Larsen J, Nguyen NL. 2004. Potentially toxic microalgae of Vietnamese waters. Opera Botanica. 140(1):5–216.
- Lim H-C, Leaw C-P, Su SN-P, Teng S-T, Usup G, Mohammad-Noor N, Lundholm N, Kotaki Y, Lim P-T. 2012. Morphology and molecular characterization of Pseudo-nitzschia (Bacillariophyceae) from Malaysian Borneo, including the new species Pseudo-nitzschia circumpora sp. Nov. J Phycol. 48(5):1232–1247.
- Park J-G, Kim E-K, Lim W-A. 2009. Potentially toxic Pseudo-nitzschia species in Tongyeong coastal waters, Korea. J Korean Soc Oceanogr. 14(3):163–170.
- Priisholm K, Moestrup Ø, Lundholm N. 2002. Taxonomic notes on the marine diatom genus Pseudo-nitzschia in the Andaman sea near the island of Phuket, Thailand, with a description of Pseudo-nitzschia micropora Sp. Nov. Diatom Res. 17(1):153–175.
- Quijano-Scheggia S, Olivos-Ortiz A, Heberto Gavino-Rodriguez J, Castro-Ochoa F, Rivera-Vilarelle M, Galicia-Perez M, Patino-Barragan M. 2011. First report of Pseudo-nitzschia brasiliana and P-micropora (Bacillariophycea) found in Cuyutlan Lagoon, Mexico. Rev Biol Mar Oceanogr. 46(2):189–197.
- Scholin CA, Gulland F, Doucette GJ, Benson S, Busman M, Chavez FP, Cordaro J, DeLong R, De Vogelaere A, Harvey J, et al. 2000. Mortality of sea lions along the central California coast linked to a toxic diatom bloom. Nature. 403(6765):80–84.
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Xu G, Li Y. 2015. Two new records of diatom genus Pseudo-nitzschia from Chinese waters and analysis of their domoic acid production. J Trop Subtrop Bot. 23(6):614–624.
- Yuan XL, Cao M, Bi GQ. 2016. The complete mitochondrial genome of Pseudo-nitzschia multiseries (Baciuariophyta). Mitochondr DNA A DNA Mapp Seq Anal. 27(4):2777–2778.
