Abstract
Talipariti tiliaceum is an evergreen mangrove associate species distributed throughout the world. In this study, the complete chloroplast genome sequence of T. tiliaceum was assembled and characterized using high-throughput sequencing data. The chloroplast genome was found to be 161 766 bp in length, consisting of large single-copy (LSC) and small single-copy (SSC) regions of 89 273 and 19 551 bp, respectively, which were separated by a pair of 26 471 bp inverted repeat (IR) regions. The genome contained 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. A phylogenetic tree including 66 chloroplast genomes from various species revealed that T. tiliaceum was most related to T. hamabo of the same genus.
Introduction
Talipariti tiliaceum (syn. Hibiscus tiliaceus), commonly known as the coast cotton tree or yellow mallow tree (Roychoudhury et al. Citation2017), is a typical mangrove associate species in tropical and subtropical coastal regions throughout the world (Abdul-Awal et al. Citation2016). The tree is fast-growing, salt- and wind-resistant, and is adapted to a wide range of mangrove environments, including inhospitable brackish swamps, waterlogged soils, and limestone-rich areas. Talipariti tiliaceum is commonly used as a traditional medicine for the treatment of fever, cough, abscesses, diarrhea, chest congestion, and ear infections (Hossain et al. Citation2015). Because of its medicinal properties, most previous research has focused on extraction methods, as well as the structural analysis and medicinal functions of its bioactive components in the leaves, flowers, and bark (Melecchi et al. Citation2002; Rosa et al. Citation2007; Li et al. Citation2008). Phylogeographical and genetic structural research based on the molecular biology of T. tiliaceum have also been reported (Tang et al. Citation2003; Takayama et al. Citation2006). However, there have been no reports on the chloroplast genome of T. tiliaceum, an important topic that would contribute to our understanding of the evolution of mangrove associate species.
Fresh young leaves of T. tiliaceum were collected from the National Nature Reserve for Mangroves in the Zhangjiang River Estuary (N23°55′, E117°26′), Yunxiao County, Fujian Province, China. Specimens were stored at the Herbarium of Taizhou University (Zhaokui Du, [email protected]) under the voucher number TZU-20200818HT01. The total genomic DNA was extracted from the leaves using a modified CTAB protocol as described by Doyle and Doyle (Citation1987). After determination of its quality and concentration, the DNA was sheared into 400-600 bp fragments by ultrasonication. An Illumina paired-end library was constructed according to the manufacturer's instructions (Illumina, San Diego, CA, USA) and paired-end sequencing was performed using Hefei Biodata Biotechnologies Inc. (Hefei, China) on the Illumina HiSq platform. The raw reads were filtered using NGS QC Toolkit v2.2 software to obtain high-quality vector- and adaptor-free reads (Patel et al. Citation2012). The filtered chloroplast genome sequences were then assembled using the program NOVOPlasty 3.6 with the T. hamabo genome (GenBank accession no. NC_030195) as a reference (Dierckxsens et al. Citation2016). Annotation of the chloroplast genome was performed using DOGMA (Wyman et al. Citation2004) and NCBI-BLAST searches.
The complete chloroplast genome of T. tiliaceum comprised 161,766 bp of double-stranded, circular DNA (GenBank accession no. MN826059), containing two IR regions of 26,471 bp, separated by LSC and SSC regions of 89,273 and 19,551 bp, respectively. The overall GC content of the genome was 36.9% and it was predicted to contain 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Six protein-coding genes (ycf1, rps7, ndhB, ycf2, rpl2, and rpl23), seven tRNA genes (trnN-GUU, trnR-ACG, trnA-UGC, trnI-GAU, trnV-GAC, trnL-CAA, and trnI-CAU), and four rRNA genes (rrn5, rrn4, rrn23, and rrn16) were duplicated in the IR regions. Nine genes contained two exons and four genes contained three exons.
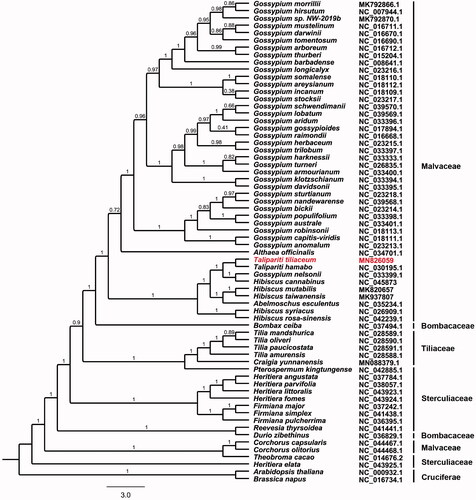
A total of 65 complete chloroplast genomes were downloaded from GenBank to investigate the phylogenetic relationships of T. tiliaceum within the genus Talipariti and other related groups. All the chloroplast genome sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013). A maximum-likelihood (ML) phylogenetic tree was constructed based on a dataset of single-copy genes that included 62 coding sequences, 25 tRNAs, and 2 rRNAs by FastTree version 2.1.10 (Price et al. Citation2010). Arabidopsis thaliana and Brassica napus were used as out-groups. The phylogenetic analysis showed that T. tiliaceum was most closely related to T. hamabo (). The sequencing of the chloroplast genome of T. tiliaceum may contribute to a better understanding of the evolution of Talipariti species within Malvaceae family.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MN826059. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA698892, SRR13610058, and SAMN17761689, respectively.
Additional information
Funding
References
- Abdul-Awal SM, Nazmir S, Nasrin S, Nurunnabi TR, Uddin SJ. 2016. Evaluation of pharmacological activity of Hibiscus tiliaceus. Springerplus. 5(1):1209–1214.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty:de novoassembly of organelle genomes from whole genome data. Nucl Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hossain H, Akbar P, Rahman S, Yeasmin S, Khan T, Rahman M, Jahan I. 2015. HPLC profiling and antioxidant properties of the ethanol extract of Hibiscus tiliaceus leaf available in Bangladesh. EJMP. 7(1):7–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li L, Sattler I, Deng Z, Groth I, Walther G, Menzel K-D, Peschel G, Grabley S, Lin W. 2008. A-seco-oleane-type triterpenes from Phomopsis sp.(strain HKI0458) isolated from the mangrove plant Hibiscus tiliaceus. Phytochemistry. 69:511–517.
- Melecchi MIS, Martinez MM, Abad FC, Zini PP, do Nascimento Filho I, Caramão EB. 2002. Chemical composition of Hibiscus tiliaceus L. flowers: a study of extraction methods. J Sep Science. 25(1–2):86–90.
- Patel RK, Mukesh J, Zhanjiang L. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2-approximately maximum-likelihood trees for large alignments. PLOS One. 5(3):e9490.
- Rosa RM, Moura DJ, Melecchi MIS, dos Santos RS, Richter MF, Camarao EB, Henriques JAP, de Paula Ramos ALL, Saffi J. 2007. Protective effects of Hibiscus tiliaceus L. methanolic extract to V79 cells against cytotoxicity and genotoxicity induced by hydrogen peroxide and tert-butyl-hydroperoxide. Toxicol In Vitro. 21(8):1442–1452.
- Roychoudhury N, Meshram P, Pandey A, Prajapati N. 2017. Hibiscus tiliaceus Linn.(family Malvaceae)–a new host plant. Indian J Forest. 40:151–154.
- Takayama K, Kajita T, Murata J, Tateishi Y. 2006. Phylogeography and genetic structure of Hibiscus tiliaceus-speciation of a pantropical plant with sea-drifted seeds. Mol Ecol. 15(10):2871–2881.
- Tang T, Zhong Y, Jian S, Shi S. 2003. Genetic diversity of Hibiscus tiliaceus (Malvaceae) in China assessed using AFLP markers. Ann Bot. 92(3):409–414.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.