Abstract
Corydalis bungeana Turcz. is a perennial herb belonging to the family Papaveraceae. Its chloroplast genome was sequenced and characterized. The cp genome of C. bungeana is 167,629 bp long with a GC content of 36.52%. A total of 144 genes were identified in this cp genome, including 79 protein-coding genes, 31 tRNAs and four rRNAs. A phylogenetic tree based on the complete nucleic acid sequence indicated that C. bungeana was classified into Corydaleae and had a close relationship with Lamprocapnos spectabilis.
Corydalis bungeana Turcz., a perennial herb, is classified into the family Papaveraceae (Chase et al. Citation2016). Previous studies have indicated that special active ingredients from dried whole plants show potential pharmacological activities, including anti-inflammatory, antibacterial and antinociceptive activities (Tian et al. Citation2019), but limited genomic and genetic resources have impeded the precise identification of C. bungeana until now. Herein, we determined the complete chloroplast genome of C. bungeana, which will provide informative data for determining the phylogeny of Corydalis and the molecular identification of C. bungeana.
Fresh leaves of C. bungeana were collected from an individual plant in a greenhouse in Chengxi District (110°19.245′ E, 19°59.757′N), Haikou, China. Fresh leaves disinfected with 75% ethyl alcohol were immediately frozen in liquid nitrogen and stored in the refrigerator at −80 °C for genomic DNA extraction. High molecular weight plant genomic DNA was extracted using a modified CTAB method (Ding et al. Citation2020) and then stored at the herbarium of Hainan Maternal and Children's Medical Center (http://www.hnwcmc.com/, Qi Wang, [email protected]) under voucher number 10_1. The pure genomic DNA was sequenced (insert size ∼350 bp) on the Illumina NovaSeq platform according to the standard protocol. Approximately 2.90 Gb raw data were obtained with a paired-end read length of 150 bp, and these raw data were filtered for high-quantity clean reads. Raw reads were deposited in the NCBI Sequence Read Archive database. The cp genome was assembled by using SPAdes v3.13.0 (Bankevich et al. Citation2012), and this draft cp genome was subsequently corrected based on GapCloser v1.12. The annotation was performed using PGA (Qu et al. Citation2019) and then submitted to GenBank (accession no. MW538958).
This cp genome of C. bungeana was 167,629 bp in length with a GC content of 36.52%, and the base composition was asymmetric (17.83% C, 17.29% G, 3012% A, and 30.91% T). This cp genome of C. bungeana was a circular molecule with a small single copy region (SSR, 18,024 bp) and a pair of inverted repeats (IR, 26,411 bp each) separated by two large single copy regions (LSC, 96,783 bp). This cp genome contained 114 genes, 79 of which were protein-coding genes, 31 were tRNA genes and four were rRNA genes. Two genes contained two introns, and nine genes had one intron, whereas 16 genes had two copies in the cp genome of C. bungeana.
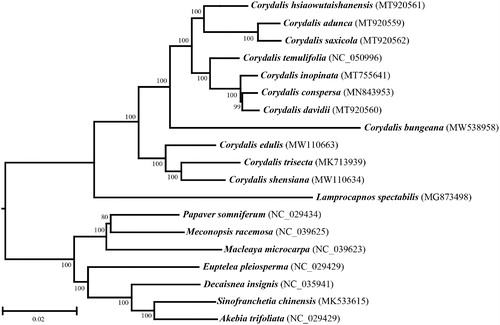
To determine its phylogenetic position within Rhoeadales, a neighbor-joining (NJ) tree was reconstructed with MEGA X (Kumar et al. Citation2018) using the complete cp genome sequence of C. bungeana and the sequences from 18 additional species from Rhoeadales (). The results suggested that C. bungeana was separated from other species of Corydalis though it was classified into the genus of Corydalis yet and the previous study with DNA barcoding also demonstrated that C. bungeana was classified into a single clade in phylogenetic tree of Corydalis based on ITS and matK (Ren et al. Citation2019). This evolutionary analysis also provided the evidence for Corydaleae had a close relationship with Lamprocapnos spectabilis from the Fumarioideae (Papaveraceae) clade (Lidén et al. Citation1997; Xu and Wang Citation2021).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the US National Center for Biotechnology Information (NCBI database) at https://www.ncbi.nlm.nih.gov/, reference number: MW538958. The associated BioProject, BioSample and SRA numbers are PRJNA692558, SAMN17348915, SRR13447699 and SSR13447700.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chase MW, Christenhusz MJM, Fay MF, et al. 2016. An update of the Angiosperm Phylogeny Group classifcation for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181(1):1–20.
- Ding X, Mei W, Lin Q, Wang H, Wang J, Peng S, Li H, Zhu J, Li W, Wang P, et al. 2020. Genome sequence of agarwood tree Aquilaria sinensis (Lour.) Spreng: the first chromosome-level draft genome in the Thymelaeceae family. GigaScience. 9(3):giaa013.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. Mega x: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lidén M, Fukuhara T, Rylander J, Oxelman B. 1997. Phylogeny and classification of Fumariaceae, with emphasis on Dicentra sl, based on the plastid gene rps16 intron. Pl Syst Evol. 206(1–4):411–420.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Ren F-M, Wang Y-W, Xu Z-C, Li Y, Xin T-Y, Zhou J-G, Qi Y-D, Wei X-P, Yao H, Song J-Y. 2019. DNA barcoding of Corydalis, the most taxonomically complicated genus of Papaveraceae. Ecol Evol. 9(4):1934–1945.
- Tian M, Yang C, Yang J, Dong H, Liu L, Ren Y, Wang Z. 2019. Ultrahigh performance liquid chromatography–electrospray ionization tandem mass spectrometry method for qualitative and quantitative analyses of constituents of Corydalis bungeana Turcz Extract. Molecules. 24(19):3463.
- Xu X, Wang D. 2021. Comparative chloroplast genomics of Corydalis Species (Papaveraceae): evolutionary perspectives on their unusual large scale rearrangements. Front Plant Sci. 11:2243.