Abstract
The complete chloroplast (cp) genome sequence of Tugarinovia mongolica Iljin has been characterized in this study. The cp genome is 153,475 bp in length, containing a large single-copy (LSC) region of 84,434 bp and a small single-copy (SSC) region of 18,715 bp, which were separated by a pair of 25,163 bp inverted repeat regions (IRs). There are 114 unique genes annotated, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. The overall GC content is 37.62%. Further, phylogenetic analysis suggested that Tugarinovia is clustered to genus Atractylodes.
Tugarinovia mongolica Iljin, included in the China Species Red List, belongs to the monotypic genus (Tugarinovia) of Asteraceae family (Fu Citation1992). The species has a limited geographical range and declining populations in the western part of Inner Mongolia. Much attention have focused on the taxonomy, origin, and biology fields (Ma et al. Citation2000; Zhao Citation2000). However, there is no complete chloroplast (cp) genome of T. mongolica in GenBank database. Here, we first reported the complete cp genome of T. mongolica, and assessed its phylogenetic position within Carduoideae.
In this study, fresh leaves of T. mongolica were sampled from the urban area of Wuhai Ordos, Inner Mongolia province, China (40°03′56″N, 106°54′08″E). Voucher specimens were deposited at the herbarium of Inner Mongolia Agricultural University (voucher number: SLBHZW202008). The genomic DNA was extracted by the modified CTAB method (Doyle Citation1987) and then sequenced using Illumina-HiSeq 2000 platform (Illumina, San Diego, CA), with a 150 bp paired-end running. NovoPlasty was used to assemble the cp genome (Dierckxsens et al. Citation2017), with the cp genome of Cynara cardunculus var. scolymus as the reference (GenBank accession no. KM035764) (Curci et al. Citation2015). Then, the assembled sequence was annotated with GeSeq (Tillich et al. Citation2017). The annotated cp genome sequence has been submitted to NCBI with an accession number of MW429288.
The complete cp genome of T. mongolica is 153,475 base pairs (bp) in length. The canonical quadripartite structure consists of a large single-copy (LSC) region of 84,434 bp, a small single-copy (SSC) region of 18,715 bp, and two inverted repeat regions (IRs) of 25,163 bp. The new sequence possesses 114 unique genes, including 80 protein-coding genes, four rRNA genes, and 30 tRNA genes. Among them, four rRNA genes (i.e. rrn16, rrn5, rrn4.5, and rrn23), seven protein-coding genes (i.e. rpl2, rpl23, ycf2, ndhB, rps7, rps12, and ycf1), and seven tRNA genes (i.e. trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU) are duplicated in the IR regions. The overall GC-content of the cp genome is 37.62%. The GC content of the LSC and SSC regions is 35.74% and 34.4%, respectively, whereas that of the IR regions is 43.1%.
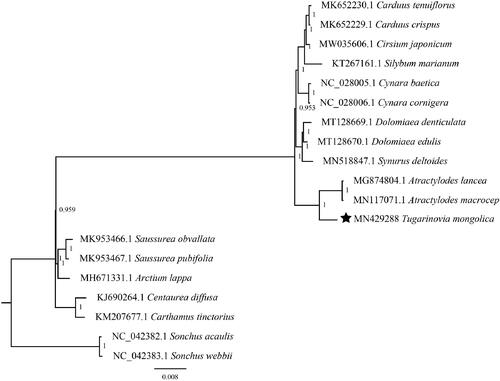
To further determine its phylogenetic position, phylogenetic analyses using 17 Carduoideae cp genomes with two Sonchus species as outgroups were performed. After alignment by MAFFT v7 (Katoh and Standley Citation2013), we found model ‘GTR + I+G’ is the fittest model for phylogenetic construction by jModelTest (Posada Citation2008). Finally, a Bayesian inference (BI) phylogenomic tree was performed in MrBayes v3.2.3 (Ronquist and Huelsenbeck Citation2003). The Markov chain Monte Carlo (MCMC) algorithm was run for 1,000,000 generations with trees sampled every 500 generations. Results in this study indicated that T. mongolica is clustered to genus Atractylodes ().
Acknowledgements
Geolocation information: The samples in this study were from the urban area of Wuhai Ordos, Inner Mongolia province, China (40°03′56″N, 106°54′08″E).
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW429288. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA700100, SAMN17824457, and SRR13649040, respectively.
Additional information
Funding
References
- Curci PL, De Paola D, Danzi D, Vendramin GG, Sonnante G. 2015. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. PLoS One. 10(3):e0120589.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Fu LG. 1992. The Red Book of Chinese plants – rare and endangered plants. Beijing: Science Press; p. 236–237.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Ma H, Wang YC, Cao R, Guo X. 2000. The embryological study of Tugarinovia mongolica. I. Megasporogenesis, microsporogenesis and development of gametophytes. Acta Bot Boreali Occid Sin. 20(3):461–466.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhao YZ. 2000. The classification and its geographycal distribution of Tugarinovia. Acta Bot Boreali Occid Sin. 20(5):873–875.