Abstract
Solanum betacea is a sub-tropical tree, and the fruit has high nutritional value, unique flavor and color. It is used as a functional ingredient in health care, food, cosmetic and pharmaceutical applications. The complete chloroplast(Cp) genome of S. betacea has been assembled and annotated in this paper. Its length was 155,937 bp, containing a large single-copy region of 86,731 bp, a small single-copy region of 18,450 bp, and a pair of IR regions of 25,378 bp in each. The complete chloroplast genome of S. betacea contained 134 genes, including 90 protein-coding genes, 36 transfer RNA genes (tRNAs), and 8 ribosome RNA genes (rRNAs). The overall GC content was 37.7% and the GC contents of the LSC, SSC, and IR regions were 35.7%, 31.8%, and 43.1%, respectively. Phylogenetic analysis with the reported chloroplast genomes revealed that S. betacea has been most closely related to Solanum torvum. These findings will provide useful information for further investigation of chloroplast genome evolution in Solanum betacea.
Solanum betacea, also known as the tree tomato, is a small Solanaceous tree or shrub, and the fruit has unique flavor and color, and is known for highly nutritious (Guo et al. Citation2007; Yang Citation2019). It is used as a functional ingredient in health care, food, cosmetic and pharmaceutical applications. (Diep et al. Citation2020). S. betacea originated in South America and now distributes in tropical and subtropical regions worldwide. In China, S. betacea has been cultivated in Yunnan province and southern Tibet as a source of fruits or vegetables (Zhang et al. Citation1998). There are no researches about the Solanum betacea chloroplast genomes. Therefore, we first assembled the complete chloroplast genome of S. betacea, which provides a genomic resource to clarify the phylogenetic relationship of this plant with other species in the Solaneae family.
Fresh Leaf samples of S. betacea were collected from Kunming, Yunnan, China (geospatial coordinates: E102°46′40″, N25°03′07″, altitude: 1954 m). The voucher specimen is deposited at the Herbarium, Kunming Institute of Botany, Chinese Academy of Sciences (specimen number: KUN 1381104). The Total DNA samples were extracted with Magnetic beads plant genomic DNA preps Kit. The DNA samples were stored at the Key Laboratory for Forest Genetic and Tree Improvement and Propagation in Universities of Yunnan Province, Southwest Forestry University, Kunming, China.
After, The obtained DNA was fragmented to construct a paired-end library with an insert size of 150 bp, and the genome sequencing was performed using Illumina Hiseq X Ten platform (Sangon Biotech (Shanghai) Co. Shanghai, China). In total, Approximately 5.5 GB of raw data genomic paired-end library of 150 bp length were obtained. The raw data (fastq files) was used to assemble the complete chloroplast genome that is automatically from scratch made by the software of GetOrganelle (Jin et al. Citation2018) with Solanum melongena as the reference, resulting in the output of a complete circle genome sequence. Genome annotation was performed with the programme Geneious R8 (Biomatters Ltd, Auckland, New Zealand), and then manually confirmed by comparison with the Solanum melongena. The CpDNA sequence of S. betacea was submitted to GenBank (accession number: MW067148).
The complete chloroplast genome of S. betacea was a circular form and length was 1,55,937 bp, which was composed of four distinct regions such as large single copy (LSC) region with a size of 86,731 bp, a small single copy (SSC) region with a size of 18,450 bp, and a pair of IRs regions of 25378 bp. The overall GC content of the genome was 37.7%, and the GC contents of the LSC, SSC, and IR regions were 35.7%, 31.8%, and 43.1%, respectively. The chloroplast genome contained a total of 134 genes, including 90 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. 6 protein-coding genes (rp12, rp123, ycf2, ndhB, rps7 and rps12), 7 tRNA genes (trnN-GUU, trnR-ACG, trnA-UGC, trnI-GAU, trnL-CAA, trnV-GAC and trnI-CAU), and all of rRNA genes (rrn4.5, rrn5, rrn16, rrn23) were duplicated in IR regions. Additionally, Among all function genes, 10 genes have a single intron, of which 6 (rpoC1, petB, rpl16, atpF, petD and rps16,) belong to protein-coding genes, and 2 (trnI-GAU, trnA-UGC) belong to tRNA genes and 5 gens (rps12, rp12, ycf3, ndhB and clpP) contain two introns.
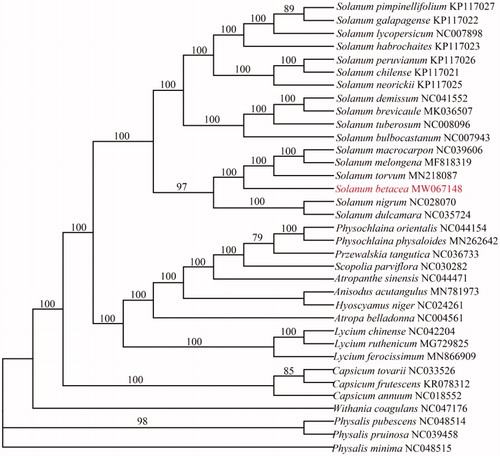
To confirm the phylogenetic position and relationship of S. betacea with other plant species in the Solaneae family, the complete chloroplast genome sequences of 13 genera (35species) were obtained from GenBank, and then aligned by MAFFT (Katoh et al. Citation2002) and trimmed properly by trimAl (Capella-Gutierrez et al. Citation2009). The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model in MEGA6.0 (Kumar et al. Citation2016). Bootstrap (BS) values were calculated from 1000 replicate analyses (). The phylogenetic tree revealed that S. betacea was closely related to Solanum torvum (). In summary, the complete chloroplast genome of S. betacea will provide useful information for molecular identification, genetic diversity research, and conservation and exploitation of S. betacea. And for phylogenetic studies and genomic resources available of Solaneae species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in [National Center for Biotechnology Information], [https://www.ncbi.nlm.nih.gov/], accession number [MW067148].
Additional information
Funding
References
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Diep TT, Rush EC, Yoo MJY. 2020. Tamarillo (Solanum betaceum Cav.): a review of physicochemical and bioactive properties and potential applications. Food Rev Int. 6:1–25.
- Guo B, Zhou W, Ye Q, Huang B. 2007. Biological characteristics and cultivation techniques of Cyphomandra betacea. Guangdong Agric Sci. 12:102–103.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assemble of a complete circular chloroplast genome using genome skimming data. BioRxiv. [in Preprint]. doi:10.1101/256479
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Yang L. 2019. Thinking on the present situation and development countermeasures of Cyphomandra betacea production in Tengchong City, Yunnan, China. South China Agric. 13(5):113–114.
- Zhang DH, Wang QP, Ma XF. 1998. Prospective tropical fruit and vegetable plant Cyphomandra betacea. Resour Dev Mark. 14:209–210.