Abstract
Dioscorea polystachya Turcz. is an important Chinese herbal medicine and the raw material of the medicine ingrediente (Chinese yam polysaccharide). It belongs to Dioscorea, which has 60 species and distribute in middle and southeast of China. In this study, we sequenced the sample of D. polystachya based on Jiaozuo, Henan and determined its complete chloroplast genome. The length of cp genome was 152,958 bp, includes two invert repeats (IR) regions of 25,492 bp, a large single-copy (LSC) region of 83,152 bp, and a short single-copy (SSC) region of 18,822 bp. There were 131 genes, which included 85 protein coding genes, 8 rRNA, and 38 tRNA, and overall GC content covered by 37.1%. Each of trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, and ndhA genes contained an intron, clpP and ycf3 contained 2 introns. The phylogenetic position showed that D. Polystachya had the closest relationship with Dioscorea alata (MG267382) and Dioscorea aspersa (NC039807).
Dioscorea polystachya Turcz. is a temperate plant that distributes in Henan, Jiangsu, Anhui, etc., and belongs to the family of Dioscoreaceae. Dioscoreaceae contains 9 genus and over 600 species around worldwide (Andres et al. Citation2017), but in China there is only 1 genus and 60 species. D. polystachya is not only used as the food and herbal medicine, but also an important raw material of the medicine ingredient (Chinese yam polysaccharide, a kind of ingredient in antihypertensive) (Luo et al. Citation2016). D. polystachya base on Jiaozuo, Henan has around 0.7–1 m long and 1.5–3 cm diameter stick root, on the root there are some dark brown or rusty flecks. It was used as raw material of medicine ingredient much more than food. We can hardly find the complete chloroplast of D. polystachya based here. So in this study, we sequenced the sample of D. Polystachya based on Jiaozuo and determined its complete chloroplast genome for the first time.
The sample of D. polystachya was collected from Jiaozuo, Henan Province (N34°55′25.525″, E113°4′30.385″) on 1 June 2020. The voucher specimen (SY20200603) was deposited in Laboratory of The fourth hospital of Changsha, Changsha (Na Hu, Email: [email protected]). We used the fresh leaves to extract total genomic DNA with the modified CTAB method (Doyle and Doyle Citation1987) and constructed the libraries with an average length of 350 bp using the NexteraXT DNA Library Preparation Kit (Illumina, San Diego, CA), then the libraries were sequenced on Illumina Novaseq 6000 platform, 2.81 Gb clean data were assembled with de novo assembler SPAdes version 3.11.0 software (St Petersburg, Russa) with default setting (Bankevich et al. Citation2012) and NOVOPlasty version 2.7.2 (Brussels, Belgium) (Dierckxsens et al. Citation2016) with kmer (K-mer = 31–33). During the assembling, the complete chloroplast of Dioscorea nipponica (MT906794) was used as reference. Finally, the assembled complete cp genome was annotated by PGA software (Qu et al. Citation2019), and submitted to GenBank under the accession number of MT712160, and SRA submitted to NCBI under the BioProject No. PRJNA694146 and SRA number: SRR13509354,
The total length of complete cp genome of D. polystachya is 152,958 bp, with a total GC content of 37.1%. The complete cp genome has a typical quadripartite structure, including a large single-copy (LSC) region of 83,152 bp, a small single-copy (SSC) region of 18,822 bp and two inverted repeat (IRs) regions of 25,492 bp. The complete cp genome contains 131 genes, including 85 protein-coding genes, 38 tRNA, and. 8 rRNA genes. trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, and ndhA genes contained an intron, and clpP and ycf3 contained 2 introns.
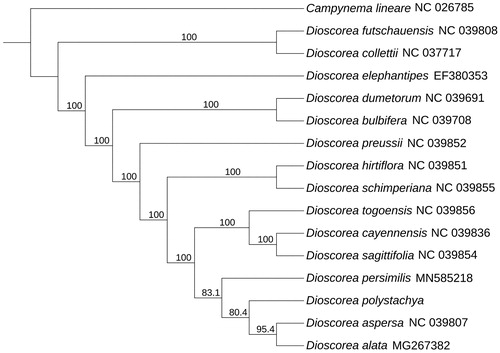
To determine the phylogenetic position of D. polystachya, the complete chloroplast genome of D. polystachya was aligned with other 15 species in Dioscorea from GenBank using Mafft-7.037 (Katoh and Standley Citation2013). Subsequently, the phylogenetic tree was constructed by IQTREE version 1.6 (Vienna, Austria) (Nguyen et al. Citation2015; Hoang et al. Citation2018) with 1000 bootstrap replicates using Best-fit model. By using Campynema lineare as out group, we got the final result. Based on the result (), we can find that D. polystachya has closely relationship with Dioscorea alata (MG267382) and Dioscorea aspersa (NC039807). This study was the first time to clearly identify the complete chloroplast of D. polystachya base on Jiaozuo and can provide useful information to further study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT712160. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA694146, SRR13509354, and SRS8097807, respectively.
References
- Andres C, Adeoluwa OO, Bhullar GS. 2017. Yam (dioscorea spp.) - sciencedirect. Encyclopedia of Applied Plant Sciences (Second Edition) 3:435–441.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Luo L, Zheng S, Huang Y, Qin T, Xing J, Niu Y, Bo R, Liu Z, Huang Y, Hu Y, et al. 2016. Preparation and characterization of Chinese yam polysaccharide PLGA nanoparticles and their immunological activity. Int J Pharm. 511(1):140–150.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch an notation of plastomes. Plant Methods. 15(1):50.