Abstract
The complete mitochondrial genome of Phintella cavaleriei is 14,325 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes, and a putative control region. The overall nucleotide composition is 35.04% A, 8.46% C, 13.41% G, and 43.09% T, with a total of A + T content of 78.13%. Ten PCGs start with typical ATN codons, two genes (cox2 and cox3) begin with TTG, and cox1 use TTA as initiation codon. Ten PCGs use usual termination codon of TAA or TAG, whereas the remaining three PCGs had an incomplete termination codon (T—). Seven tRNAs (trnY, trnC, trnG, trnN, trnH, trnP, and trnV) lacked the TΨC arm stem, while two tRNAs (trnS1 and trnS2) lost the dihydrouracil (DHU) arm. Phylogenetic analysis based on 13 PCGs indicated that P. cavaleriei was closely related to Cheliceroides longipalpis, and clustered within Salticidae clade.
Keywords:
The jumping spider Phintella cavaleriei belongs to the family of Salticidae, which includes 6329 described species in 659 genera (Wheeler et al. Citation2017; Kanesharatnam and Benjamin Citation2019; World Spider Catalog Citation2021). Phintella cavaleriei is an important predator of many agricultural pests and mainly distributed in China and Korea (Cui et al. Citation2012; Kim and Lee Citation2014). In this study, adult individuals of P. cavaleriei were collected from organic tobacco fields in Enshi City, Hubei Province, China (N30°20i, E109°25E). Samples were preserved in 95% ethanol and stored in the Natural Enemy Insect Specimen Room of Enshi Tobacco Company under the voucher number ETC-2020-01. Genomic DNA was extracted from whole body of a single specimen using the EasyPure® Genomic DNA Kit (TransGen, Beijing, China). The complete mitogenome sequence of P. cavaleriei was amplified by polymerase chain reaction (PCR) with the LA PCR™ Kit (TaKaRa Bio Inc, Dalian, China). Initially, partial sequences of cox1, cox3, nd1, nd5, 12S rRNA, and 16S rRNA were amplified by using the universal primers, which were designed on the basis of the conserved regions of 10 arachnida mitogenomes. These fragments were then used for designing six specifc primers to amplify the remaining mitogenomic sequences in several PCR steps.
The complete mitogenome of P. cavaleriei (GenBank accession number MW540530) is a typical circular DNA molecule of 14,325 bp in length, and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (12S rRNA and 16S rRNA), and a putative control region (Boore Citation1999). The gene order and orientation of P. cavaleriei are identical with other spider mitogenomes (Pan et al. Citation2016; Wang et al. Citation2016). The overall base composition of P. cavaleriei mitogenome is A (35.04%), C (8.46%), G (13.41%), and T (43.09%), with a total of A + T content of 78.13%. The AT-skew and GC-skew of this genome were −0.103 and 0.226, respectively. Gene overlaps were found in 22 locations and their total length was 302 bp. The longest overlap was 35 bp in length and resided between nad4 and nad4L. There were 6 intergenic spacer regions comprising a total of 122 bp and the largest spacer (70 bp) resided between trnA and trnN. The length of 22 tRNAs ranged from 49 bp (trnC) to 80 bp (trnM), A + T content ranged from 69.81% (trnN) to 87.93% (trnT). Nine tRNAs lacked the potential to form the cloverleaf-shaped secondary structure. Seven of them (trnY, trnC, trnG, trnN, trnH, trnP, and trnV) lacked the TΨC arm stem, two tRNAs (trnS1 and trnS2) lost the dihydrouracil (DHU) arm. The 16S rRNA (1036 bp) was located between trnL1 and trnV, and 12S rRNA (692 bp) resided between trnV and trnQ, and their A + T contents were 83.01% and 81.36%, respectively. The control region was located between trnQ and trnM genes with a length of 663 bp, and the A + T content was 79.03%.
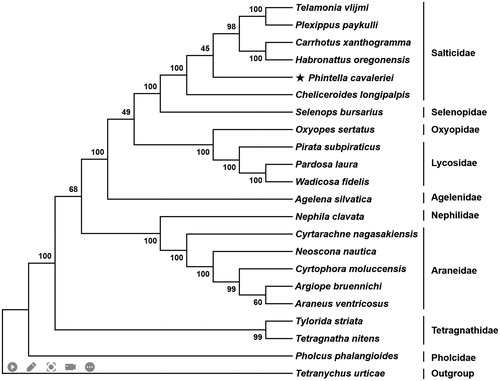
Among the 13 PCGs, nad1, nad4, nad4L, and nad5 were encoded on the light strand (L-strand), while the remaining nine genes were encoded on the heavy strand (H-strand). The A + T content of these 13 PCGs ranged from 72.01% (cox1) to 88.24% (atp8). The cox1 initiated with TTA as the start codon, cox2 and cox3 started with TTG, atp8, nad2, nad4L, nad5, and nad6 started with ATT, and the remaining five PCGs (atp6, cob, nad1, nad4, and nad6) started with ATA. Ten PCGs terminated with conventional stop codons (TAA and TAG), while nad2, nad4, and cob used incomplete codon (T—) as termination codon. Based on the concatenated amino acid sequences of 13 PCGs, the neighbor-joining method was used to construct the phylogenetic relationship of P. cavaleriei with 20 other spiders by MEGA7 (Kumar et al. Citation2016). The result showed that P. cavaleriei was closely related to Cheliceroides longipalpis, and clustered within Salticidae clade ().
Figure 1. Phylogenetic tree showing the relationship between Phintella cavaleriei and 20 other representative spiders based on neighbor-joining method. GenBank accession numbers used in the study are the following: Agelena silvatica (KX290739), Araneus ventricosus (NC_025634), Argiope bruennichi (NC_024281), Carrhotus xanthogramma (NC_005942), Cheliceroides longipalpis (MH891570), Cyrtarachne nagasakiensis (KR259802), Cyrtophora moluccensis (KM820884), Habronattus oregonensis (AY571145), Neoscona nautica (KR259804), Nephila clavata (NC_008063), Oxyopes sertatus (KM272950), Pardosa laura (KM272948), Phintella cavaleriei (MW540530), Pholcus phalangioides (NC_020324), Pirata subpiraticus (NC_025523), Plexippus paykulli (NC_024877), Selenops bursarius (NC_024878), Telamonia vlijmi (NC_024287), Tetragnatha nitens (KP306790), Tetrancychus urticae (EU345430.1), Tylorida striata (MN615900), and Wadicosa fidelis (NC_026123). T. urticae was used as an outgroup. Spiders determined in this study was marked with an asterisk.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number MW540530.
Additional information
Funding
References
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Cui HQ, Zhang Q, Hou WH, Li WD, Dai JH, Yuan CX. 2012. Survey on Spider Species in Tea Garden in Enshi. Hubei Agr Sci. 51:2735–2737.
- Kanesharatnam N, Benjamin SP. 2019. Multilocus genetic and morphological phylogenetic analysis reveals a radiation of shiny South Asian jumping spiders (Araneae, Salticidae). Zookeys. 839:1–81.
- Kim ST, Lee SY. 2014. Arthropoda: Arachnida: Araneae: Clubionidae, Corinnidae, Salticidae, Segestriidae. Spiders. Invertebr Fauna Korea. 21:1–186.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Pan WJ, Fang HY, Zhang P, Pan HC. 2016. The complete mitochondrial genome of pantropical jumping spider Plexippus paykulli (Araneae: Salticidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1490–1491.
- Wang ZL, Li C, Fang WY, Yu XP. 2016. The complete mitochondrial genome of orb-weaving spider Araneus ventricosus (Araneae: Araneidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):1926–1927.
- Wheeler WC, Coddington JA, Crowley LM, Dimitrov D, Goloboff PA, Griswold CE, Hormiga G, Prendini L, Ramírez MJ, Sierwald P, Almeida‐Silva L, et al. 2017. The spider tree of life: phylogeny of Araneae based on target‐gene analyses from an extensive taxon sampling. Cladistics. 33(6):574–616.
- World Spider Catalog. 2021. Version 22.0. Natural History Museum Bern. [cited 2020 Jul 9]. Available at https://wsc.nmbe.ch/
