Abstract
Ferula sinkiangensis K. M. Shen is a valuable traditional Chinese medicine historically used to treat stomachache and rheumatoid arthritis. The chloroplast genome of Ferula genus plant has not been previously reported. This study reported the complete chloroplast genome sequence of F. sinkiangensis based on high-throughput sequencing. The genome was 166,583 bp in length, containing a small single-copy (SSC) region of 17,595 bp and a large single-copy (LSC) region of 85,242 bp, separated by two inverted repeats (IRs) of 31,873 bp, each. The genome contained 114 unique genes, including 80 protein-coding genes (PCGs), four rRNA genes, and 30 tRNA genes. In addition, 17 genes contained one or two introns, including nine PCG genes with a single intron, two PCG genes harboring two introns, and six tRNA genes harboring a single intron. In this study, F. sinkiangensis K. M. had the closest genetic relationship with Torilis scabra and clustered with the Umbelliferae family species.
The Ferula resin is used globally as a traditional medicinal herb, and Chinese Pharmacopoeia recently documented Ferula sinkiangensis as one of the main primitive plant. The resin of F. sinkiangensis is used to treat many diseases, especially stomachache, flatulence, and poor digestion (Iranshahy and Iranshahi Citation2011; Mahendra and Bisht Citation2012). Modern pharmacology and biological studies have shown that F. sinkiangensis has antiviral (Nazari and Iranshahi Citation2011), antifungal (Kavoosi et al. Citation2013), cancer chemopreventive, and anti-influenza effects (Iranshahi et al. Citation2010; Zhou et al. Citation2017).
In China, the Ferula genus is mostly distributed in Xinjiang but has been destroyed due to excessive collection. In addition, slow growth, change in reclamation, irrigation, road construction, and deterioration of the native habitat contribute to the annual shrinkage of Ferula resources. Currently, wild resources of F. sinkiangensis are extremely endangered; thus, many researchers are interested in studying its endangerment mechanism to protect and utilize it. This study assembled the complete chloroplast genome of this species by Illumina sequencing (San Diego, CA) to facilitate its phylogenetic analysis, conservation genetics, and ensure sustainable utilization. The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession MW411057. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA715948, SUB9326528, and SAMN18388803, respectively.
The number of species in the Umbelliferae family with published chloroplast genomes is relatively small. In this study, the chloroplast genome of a Ferula genus plant was assembled for the first time. The determination of this complete plastid genome sequence is a useful resource and provides molecular data to illuminate the F. sinkiangensis evolution and its endangerment mechanism.
In this study, fresh leaves of F. sinkiangensis were collected from Baishidun Township of Yining City (82.0574 E, 43.6743 N), Xinjiang, China. The voucher specimen (654021120525001) was deposited at the Xinjiang Institute of Chinese Materia Medica and Ethnical Materia. The genomic DNA was extracted using the DNeasy Plant Mini Kit (QIAGEN, Valencia, CA). High-throughput sequencing was conducted using the Illumina HiSeq X Ten platform, and data processing was according to Yan et al. (Citation2019).
The complete chloroplast genome was annotated using Torilis scabra (MN105615) as a reference. The chloroplast genome of F. sinkiangensis has a circular quadripartite structure which is similar to major angiosperm chloroplast genomes (Hahn C et al. Citation2013). It has a length of 166,583 bp, contained a small single-copy (SSC) region of 17,595 bp and a large single-copy (LSC) region of 85,242 bp separated by two inverted repeats (IRs) of 31,873 bp, each. Similar to the chloroplast genomes of other higher plants, the base composition is asymmetric (30.7% A, 19% C, 18.9% G, and 31.4% T) with an overall GC content of 62.1%.
The F. sinkiangensis chloroplast genome contained 114 unique genes, including 80 protein-coding genes (PCGs), four rRNA genes, and 30 tRNA genes. In addition, 17 genes contain one or two introns, which include nine PCG genes (atpF, ndhA, ndhB, petB, rpl2, rpl16, rpoC1, rps12, rps16) possessing a single intron, two PCG genes (clpP, ycf3) harboring two introns, and six tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) harboring a single intron.
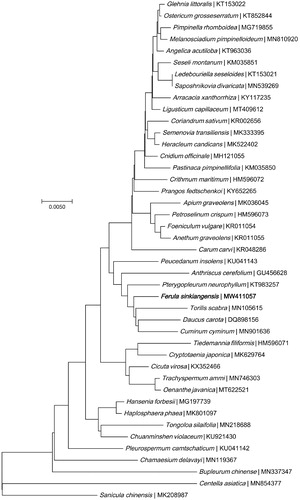
A neighbor-joining phylogeny was constructed using MEGA7.0 (Kumar et al. Citation2016) based on nine genes (accD, atpA, atpB, matK, psbB, psbD, rbcL, rpoA, rpoC1) in the chloroplast genome of F. sinkiangensis and 42 other species from the Umbelliferae family. The phylogenetic tree analysis showed that F. sinkiangensis was closely related to the T. scabra ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
We confirm that the data supporting the findings of this study are available within the article and its supplementary materials. The data that support the study findings have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/) under accession number MW411057.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—abaiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Iranshahi M, Sahebkar A, Hosseini ST, Takasaki M, Konoshima T, Tokuda H. 2010. Cancer chemopreventive activity of diversin from Ferula diversivittata in vitro and in vivo. Phytomedicine. 17(3–4):269–273.
- Iranshahy M, Iranshahi M. 2011. Traditional uses, phytochemistry and pharmacology of asafoetida (Ferula assafoetida oleo-gum-resin)—a review. J Ethnopharmacol. 134(1):1–10.
- Kavoosi G, Tafsiry A, Ebdam AA, Rowshan V. 2013. Evaluation of antioxidant and antimicrobial activities of essential oils from Carum copticum Seed and Ferula assafoetida Latex. J Food Sci. 78(2):T356–T361.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Mahendra P, Bisht S. 2012. Ferula asafoetida: traditional uses and pharmacological activity. Pharmacogn Rev. 6(12):141–146.
- Nazari ZE, Iranshahi M. 2011. Biologically active sesquiterpene coumarins from Ferula species. Phytother Res. 25(3):315–323.
- Yan F, Tao X, Wang Q-L, Juan ZY, Zhang C-M, Yu HL. 2019. The complete chloroplast genome sequence of the medicinal shrub Daphne giraldii Nitsche. (Thymelaeaceae). Mitochondrial DNA Part B. 4(2):2685–2686.
- Zhou Y-T, Fang X, Zhang G-Q, Qu H-X, Yang D-S, Han X-Q. 2017. Recent advances on bioactive constituents in Ferula. Drug Dev Res. 78(7):321–331.