Abstract
We assembled and annotated the complete mitochondrial genome of Scatoglyphus polytrematus. It is the first complete mitochondrial genome sequence from the genus Scatoglyphus. The mitogenome was 13,966 bp in length and contains 37 genes (including 13 protein-coding genes, 22 transfer RNA (tRNA), and two ribosomal RNA (rRNA)), and one largest non-coding region. The gene arrangement of S. polytrematus is consistent with the pattern of possible common ancestor of astigmatid mites. In the present study, phylogenetic analysis shows that genus Scatoglyphus was clustered into one branch with other Acaridae species.
Mites of the family Acaridae are economically important polyphagous pest commonly living on stored products and also responsible for allergic reactions to humans (Cui Citation2014). Scatoglyphus polytrematus (Berlese 1913) belongs to Astigmatina, Acaridae, which was the only species reported in genus Scatoglyphus. To date, six mitogenomes from species of Acaridae have been sequenced. Three mitochondrial transfer RNA (tRNA) genes (trnF, trnS1, and trnQ) were reported as lost in Tyrophagus longior (Yang and Li Citation2015). Here, we present the complete mitogenome of S. polytrematus, analyze its composition.
We collected samples of S. polytrematus from piles of firewood in Wuhu, southeast China (118°38′E, 31°33′N), in July 2019. Voucher specimen was deposited in the herbarium of Department of Health Inspection and Quarantine, Wannan Medical College (Entao Sun, [email protected]) (under the accession number WNMC0820190410). Mites were stored in 100% ethanol at −20 °C until use. The whole-genomic DNA was extracted by standard phenol–chloroform extraction (Zhang and Alvarado Citation2018). Sequencing libraries were prepared by Shanghai BIOZERON Company (Shanghai, China) and sequenced on the Illumina Hiseq 4000 (San Diego, CA). The assembled genome was annotated using the MITOS WebServer (Bernt et al. Citation2013). The PCGs boundaries were confirmed manually by MEGA X software (Kumar et al. Citation2018), and BLASTp (Altschul et al. Citation1997). We annotated tRNAs using ARWEN (Laslett and Canback Citation2008), tRNAscan-SE (Schattner Citation2005), and manual identification based on the anticodon and predicted secondary structure.
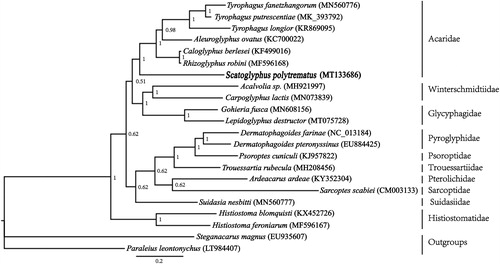
Figure 1. Phylogenetic tree inferred from mitochondrial genome sequences using Bayesian methods. Branch lengths presented here follow the Bayesian methods analysis. Node numbers indicate Bayesian posterior probabilities (BPPs).
The complete mitogenome of S. polytrematus (GenBank: MT133686) is 13,966 bp. The overall base composition of the entire S. polytrematus mitogenome consisted of 24.1% A, 46.1% T, 10.2% C, and 19.6% G, resulting in a negative AT-skew (-0.3125) and a positive GC-skew (0.3122). The genome contains 37 genes, including 13 protein-coding genes (PCGs), 22 tRNA genes, two ribosomal RNA (rRNA) genes, and one D-loop. The gene arrangement of S. polytrematus is consistent with the most available astigmatid mites, which is supposed to reflect the possible common ancestor of astigmatid mites (Li and Xue Citation2019). The length of the tRNAs ranged from 47 to 62 bp. Only the trnK showed the typical cloverleaf. Other tRNAs showed the reduction of tRNA-D- and/or T-arms, like those found in other astigmatid mites.
To infer the phylogenetic position of S. polytrematus within the Astigmatina, we generated a data set of 22 mite taxa (20 astigmatid mites and two oribatid mites), including the nucleotide sequences and amino acid sequences of the 13 PCGs. The nucleotide and amino acid sequences of the PCGs were aligned separately using the TranslatorX server (Abascal Citation2010), where MAFFT is used to build the protein alignment (Katoh and Standley Citation2013). For the nucleotide sequences, translation was done under the invertebrate mitochondrial genetic code. The large gaps and ambiguous sites were deleted by Gblocks v.0.91b (Castresana Citation2000). Phylogeneticanalyses were conducted using Bayesian inference (BI) (Ronquist et al. Citation2012) method. The phylogenetic analysis supported the monophyly of Acaridae (). The genus Scatoglyphus was placed under family Acaroidae, which is congruent with the current classification systems (Krantz and Walter Citation2009).
The complete mitogenome of S. polytrematus was determined in this study. This information from our study has important ramifications for understanding of mitogenome evolution in astigmatid mites.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MT133686. Associated BioProject and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/608033 and SAMN14116537, respectively.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–W13.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Cui YB. 2014. When mites attack: domestic mites are not just allergens. Parasit Vectors. 7(1):411.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Krantz GW, Walter DE. 2009. A manual of acarology. Lubbock (TX): Texas Tech University Press.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li WN, Xue XF. 2019. Mitochondrial genome reorganization provides insights into the relationship between oribatid mites and astigmatid mites (Acari: Sarcoptiformes: Oribatida). Zool J Linn Soc. 187(3):585–598.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Yang BH, Li CP. 2015. Characterization of the complete mitochondrial genome of the storage mite pest Tyrophagus longior (Gervais) (Acari: Acaridae) and comparative mitogenomic analysis of four acarid mites. Gene. 576:807–819.
- Zhang SS, Alvarado AS. 2018. Planarian high molecular weight DNA isolation by spooling. Methods Mol Biol. 1774:277–284.
