Abstract
This paper reports on the complete mitochondrial (mt) genome of a horseshoe crab, Tachypleus gigas (T. gigas), in Kuala Kemaman, Terengganu, Malaysia. Whole-genome sequencing of hemocyte DNA was performed with Illumina HiSeq system and the generated reads were de novo assembled with ABySS 2.1.5 and reassembled using mitoZ against Carcinoscorpius rotundicauda and Limulus polyphemus, resulting in a contig of 15 Kb. Phylogenetic analysis of the assembled mt genome suggests that the Tachypleus gigas is closely related to Tachypleus tridentatus than to Carcinoscorpius rotundicauda.
Introduction
Horseshoe crabs (HSCs) are important species in selected marine ecosystems, being the source of protein for other marine species and contributing as bioturbators to the rearrangement and aeration of marine sediments. HSCs also have an important role in the biomedical and pharmaceutical industries, and an important subject of research from the biological, evolutionary, and ecological standpoints (Manca et al. Citation2017; Krisfalusi-Gannon et al. Citation2018). A combination of factors such as environmental changes and uncontrolled economical exploitation threaten the survival of HSCs which encourages the genetic research on these species to better understand their mechanisms of adaptation, genetic diversity, and immune defensive system.
HSCs are marine, brackish water arthropods of the phylum Arthropoda, subphylum Chelicerata, order Xiphosura, suborder Xiphosurida, family Limulidae, and subfamily Tachypleinae. Based on their unique and limited morphological evolution, HSCs are considered as ‘living fossils.’ The evolutionary history of HSCs dates around 450 million years ago (Zhou et al. Citation2020). HSCs present some ordinary traits of crustaceans but are more associated with arachnids such as spiders and scorpions (Chen et al. Citation2016). At present, there are only four known HSC species (Gong et al. Citation2019). The Carcinoscorpius rotundicauda (C. rotundicauda) (mangrove) species is distributed across South and Southeast Asia, mainly in Thailand (Kanchanapongkul Citation2008), Malaysia, Eastern India, Bangladesh, Cambodia, southern and northern Vietnam, southern China, Sumatra, Java, and Borneo (Vestbo et al. Citation2018); Limulus polyphemus (L. polyphemus) (Atlantic or American) lives in the Gulf of Mexico and in the American Atlantic coast (Zhu et al. Citation2020); Tachypleus tridentatus (T. tridentatus) (Chinese, Japanese or tri-spine) is distributed in Southeast and East Asia mainly in Sabah, Malaysia (Mohamad et al. Citation2016), southern Japan, China, Taiwan, northern Vietnam, Philippines, Island of Borneo and Java (Vestbo et al. Citation2018) and Tachypleus gigas (T. gigas) (Indo-Pacific, Indonesian, Indian or southern), which is found in South and Southeast Asia in places such as Malaysia, in the States of Pahang (Razak et al. Citation2017) and Terengganu (Rozihan and Ismail Citation2012), as well as in Thailand and India (Vestbo et al. Citation2018).
Genomes and transcriptomes from different HSC species (L. polyphemus, T. tridentatus, and C. rotundicauda) have been described previously (Ding et al. Citation2005; Battelle et al. Citation2016; Chen et al. Citation2016; Chesmore et al. Citation2016; Gong et al. Citation2019; Liao et al. Citation2019; Lou et al. Citation2020; Shingate et al. Citation2020; Zhou et al. Citation2020). However, information about the genome of T. gigas has been scarce, which has limited the knowledge on the biology of this species. In this regard, a recent study reported the genome sequence of a T. gigas specimen from Singapore (Shingate et al. Citation2020). Our current study, therefore, complements this recent report by providing the mitochondrial (mt) genome data of a specimen of the same species from a different geographical habitat.
The study of the mt DNA sequence of different HSCs species has been a focus of scientific interest due to its importance to establish their evolutionary and phylogenetic relationship (Baek et al. Citation2014). The mt DNA sequence of L. polyphemus, C. rotundicauda, and T. tridentatus have been reported previously (Lavrov et al. Citation2000; Baek et al. Citation2014). In this study, we reported the mt DNA sequence of a T. gigas specimen from Terengganu, Malaysia, whose genome has been sequenced by our group, together with a phylogenetic study based on the mt protein-coding genes which will contribute to the better understanding of genomic features of this endangered species.
Materials and methods
Sample collection
An adult HSC was purchased from the fishermen who collected it from the coastal area in Kuala Kemaman, Terengganu, Malaysia. The HSC was transported to the Universiti Malaysia Terengganu (UMT) Hatchery at night to avoid direct heat from the environment. HSCs are sensitive to heat and this would induce stress. The HSC was allowed to acclimatize for 3 days and was cleaned thoroughly before sample collection. The hemolymph was collected using pyrogen-free apparatus in sterile condition in a Biological Safety Cabinet Class II (ESCO, USA) following patented techniques (MY-155541-A) from Makmal Belangkas, UMT. Hemolymph (2 mL) was drawn out and the HSC was transported back to the sea after sample extraction.
DNA purification
DNA from the hemocytes was extracted using a DNA purification kit (QIAamp DNA Blood Mini Kit, Qiagen) according to the manufacturer’s instructions. Briefly, 1 mL of fresh hemolymph was added to 40 ml of 3% NaCl contained in 50 mL sterile-polypropylene-centrifuge tubes and mixed thoroughly. The tube was centrifuged at 2000 × g for 5 min, then the supernatant was discarded. The genomic DNA was extracted from the cell pellet. DNA integrity and quantification were determined by agarose gel electrophoresis, Nanodrop and Qubit™ (Invitrogen).
Library construction, quality control, and sequencing
A total amount of 500 ng DNA was used as input material for the sample preparation. Sequencing libraries were generated using NEBNext® Ultra II DNA Library Prep Kit (New England Biolabs, England) according to the manufacturer's recommendations and index sequences were added to the genomic libraries. The genomic DNA was randomly fragmented to a size of 350 bp by Covaris cracker (Covaris, USA), before the DNA fragments were end-polished, A-tailed, and ligated with the full-length adapter for Illumina sequencing with further PCR amplification. The PCR products were purified (AMPure XP system) and the libraries were analyzed for size distribution by Agilent 2100 Bioanalyzer and quantified using real-time PCR. The qualified libraries were loaded into Illumina HiSeq sequencer after pooling according to its effective concentration and expected data volume.
Quality control and mitochondrial assembly
Paired-end Illumina sequences were first removed of sequence adaptors and reads with low-quality scores using bbduk of the BBTools Packages (https://jgi.doe.gov/data-and-tools/bbtools/). Reads were assembled de novo using ABySS 2.1.5 (Jackman et al. Citation2017). To determine the presence of mt genome in the Abyss kmer72 assembly, the mt genome of C. rotundicauda (NC_019623) was used to search against the kmer72 assembly using blastn. The search recovered a contig of approximately 14 Kb, but this contig is linear and less than the size of the reference mt genome of approximately 15 Kb. The Abyss contigs were used as input for reassembly with mitoZ (Meng et al. Citation2019) by comparing to C. rotundicauda and L. polyphemus (NC_003057.1). The open reading frames were predicted with reference to the invertebrate mt translational table, and annotation was done using MITOS1 software (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). A pairwise comparison of mitogenome sequences between T. gigas and C. rotundicauda was performed using BLASTN 2.9.0.
Phylogenetic analysis
Mt protein-coding genes nucleotide sequences from different organisms: Ixodes hexagonus (AF081828), T. tridentatus (JQ739210 and FJ860267), T. gigas (MT241669), C. rotundicauda (JX437074 and JQ178358) and L. polyphemus (NC_003057 and JX983598), were selected to determine the phylogenetic analysis of T. gigas.
The nucleotide sequence alignment was performed using ClustalW software and a maximum likelihood tree was constructed using MEGA X with 1000 bootstrapping (Kumar et al. Citation2018).
Results and discussion
Mitochondria genome
The sequencing reads first de novo assembled using Abyss and the Abyss contigs were then used as input for the mitoZ (Meng et al. Citation2019) reassembly against C. rotundicauda and L. polyphemus. The final assembled contig result showed a genome size of 15 Kb and exhibited similar gene organization compared to C. rotundicauda and L. polyphemus. Gene annotation with MITOS1 showed the mitochondrial genome contained 13 protein-coding genes (Supplementary Figure 1).
Phylogenetic analysis
Several phylogenetic studies have been focused on HSCs, however, the relationship between L. polyphemus and the Asian HSC species is not well defined. A fragment of mt cox1 (or COI) gene, which is a ‘DNA barcode’ used as a universal marker for species identification (Pentinsaari et al. Citation2016), either alone or in combination with other markers had been used for HSC phylogenetic studies (Avise et al. Citation1994; Kamaruzzaman et al. Citation2011; Obst et al. Citation2012; Baek et al. Citation2014; Dhar et al. Citation2016; John et al. Citation2016). HSC phylogenetic studies using other markers have also been reported (Sekiguchi and Sugita Citation1980; Shishikura et al. Citation1982; Miyata et al. Citation1984; Iwanaga and Kawabata Citation1998; Ismail and Sarijan Citation2011; Lamsdell Citation2016).
In the present study, the phylogenetic position of T. gigas among the HSC species was analyzed using whole mt protein-coding genes with L. hexagonus as the outgroup. T. gigas in this study was clustered together with the other T. gigas sequence (MT241669) and showed a closer relationship with T. tridentatus than with C. rotundicauda (). Our findings are consistent with previous phylogenetic reports (Obst et al. Citation2012; Dhar et al. Citation2016; John et al. Citation2016; Lamsdell Citation2016). A recent study using a genome-wide protein dataset supports the Tachypleus monophyly (Shingate et al. Citation2020).
Figure 1. Comparative phylogenetic tree of Tachypleus gigas (Class: Merostomata; Family: Limulidae). Maximum likelihood tree of mitochondrial-protein coding genes using MEGA X with 1000 bootstrapping. Ixodes hexagonus (AF081828) was used as an out group. Red arrow: Mitochondrial DNA sequence (MW446184) obtained in this study.
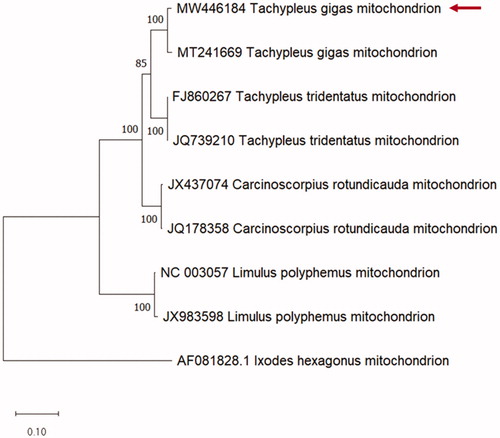
However, in contrast to our results, Avise et al. (Citation1994) and Kamaruzzaman et al. (Citation2011) showed a closer relation of T. gigas with C. rotundicauda than with T. tridentatus.
Iwanaga and Kawabata (Citation1998) and Shishikura et al. (Citation1982) conducted phylogenetic studies based on coagulogen and peptide C, respectively, and a closer relationship between T. tridentatus and C. rotundicauda compared to T. gigas was reported.
Baek et al. (Citation2014) reported a phylogenetic study based on whole mt genomes, which did not include T. gigas in their analysis.
Our study found, after pairwise mitogenome similarity comparison, mt DNA sequence differences (97% of identity) compared to the mt DNA sequence reported by Shingate et al. (Citation2020), which could be related to the influence of the different geographic ecosystems. The specimen studied by Shingate et al was obtained from East Coast Park, Singapore, while our specimen was collected from Kuala Kemaman, Terengganu, Malaysia.
Genetic differences have been found in the same HSC species, including T. gigas, belonging to different geographical locations, which could be explained by multiple factors such as the limited migratory capacity of HSCs that influence the gene flow between populations, among others (Xia Citation2000; Ismail and Sarijan Citation2011; Rozihan and Ismail Citation2011; Obst et al. Citation2012; Adibah et al. Citation2015; Liew et al. Citation2015; Periasamy et al. Citation2017).
In summary, this study presents the whole mt DNA sequence of a T. gigas specimen from Terengganu, Malaysia, and suggests a close phylogenetic relationship between T. gigas and T. tridentatus, compared to C. rotundicauda.
Supplemental Material
Download MS Word (103 KB)Acknowledgment
We thank the Universiti Malaysia Terengganu (UMT) Hatchery for providing necessary facilities for the study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI Genbank database at (https://www.ncbi.nlm.nih.gov) with the accession number (MW446184).
Additional information
Funding
References
- Adibah AB, Ng WL, Tan SG. 2015. The Malay Peninsula as a barrier to gene flow in an Asian horseshoe crab species, Carcinoscorpius rotundicauda Latreille. Biochem Syst Ecol. 60:204–210.
- Avise JC, Nelson WS, Sugita H. 1994. A speciational history of “living fossils”: molecular evolutionary patterns in horseshoe crabs. Evolution. 48(6):1986–2001.
- Baek SY, Choi EH, Jang KH, Ryu SH, Park SM, Suk HY, Chang CY, Hwang UW. 2014. Complete mitochondrial genomes of Carcinoscorpius rotundicauda and Tachypleus tridentatus (Xiphosura, Arthropoda) and implications for chelicerate phylogenetic studies. Int J Biol Sci. 10(5):479–489.
- Battelle B-A, Ryan JF, Kempler KE, Saraf SR, Marten CE, Warren WC, Minx PJ, Montague MJ, Green PJ, Schmidt SA, et al. 2016. Opsin repertoire and expression patterns in horseshoe crabs: evidence from the genome of Limulus polyphemus (Arthropoda: Chelicerata). Genome Biol Evol. 8(5):1571–1589.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen M, Wang C, Wang W, Ji G, Hu B, Du M, Liu G, Li Z, Wang W, Lin X, et al. 2016. De novo assembly and characterization of early embryonic transcriptome of the horseshoe crab Tachypleus tridentatus. PloS One. 11(1):e0145825.
- Chesmore KN, Watson WH, Chabot CC. 2016. Identification of putative circadian clock genes in the American horseshoe crab, Limulus polyphemus. Comp Biochem Physiol Part D Genom Proteom. 19:45–61.
- Dhar B, Ghose A, Kundu S, Malvika S, Devi NN, Choudhury A, Ghorai S, Trivedi S, Ghosh SK. 2016. DNA barcoding: Molecular positioning of living fossils (horseshoe crab). In: Trivedi S, Ansari AA, Ghosh SK, Rehman H, editors. DNA barcoding in marine perspectives. Amsterdam (The Netherlands): Springer; p. 181–199.
- Ding JL, Tan KC, Thangamani S, Kusuma N, Seow WK, Bui THH, Wang J, Ho B. 2005. Spatial and temporal coordination of expression of immune response genes during Pseudomonas infection of horseshoe crab, Carcinoscorpius rotundicauda. Genes Immun. 6(7):557–574.
- Gong L, Fan G, Ren Y, Chen Y, Qiu Q, Liu L, Qin Y, Liu B, Jiang L, Li H, et al. 2019. Chromosomal level reference genome of Tachypleus tridentatus provides insights into evolution and adaptation of horseshoe crabs. Mol Ecol Resour. 19(3):744–756.
- Ismail N, Sarijan S. 2011. Phylogenetic inference from 18S rRNA gene sequences of horseshoe crabs, Tachypleus gigas between Tanjung Dawai, Kedah and Cherating, Pahang, Peninsular Malaysia. Int J Agric Biol Sci. 1:95–98.
- Iwanaga S, Kawabata S. 1998. Evolution and phylogeny of defense molecules associated with innate immunity in horseshoe crab. Front Biosci. 3(4):D973–D984.
- Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, et al. 2017. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27(5):768–777.
- John BA, Sheikh HI, Jalal K, Zaleha K, Kamaruzzaman B. 2016. Revised phylogeny of extant xiphosurans (horseshoe crabs). In: Trivedi S, Ansari AA, Ghosh SK, Rehman H, editors. DNA barcoding in marine perspectives. Amsterdam (The Netherlands): Springer; p. 113–130.
- Kamaruzzaman BY, John BA, Zaleha K, Jalal KCA. 2011. Molecular phylogeny of horseshoe crab. Asian J Biotechnol. 3(3):302–309.
- Kanchanapongkul J. 2008. Tetrodotoxin poisoning following ingestion of the toxic eggs of the horseshoe crab Carcinoscorpius rotundicauda, a case series from 1994 through 2006. Southeast Asian J Trop Med Public Health. 39(2):303–306.
- Krisfalusi-Gannon J, Ali W, Dellinger K, Robertson L, Brady TE, Goddard MK. 2018. The role of horseshoe crabs in the biomedical industry and recent trends impacting species sustainability. Front Mar Sci. 5:185.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lamsdell JC. 2016. Horseshoe crab phylogeny and independent colonizations of fresh water: ecological invasion as a driver for morphological innovation. Palaeontology. 59(2):181–194.
- Lavrov DV, Boor JL, Brown WM. 2000. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol Biol E. 17(5):813–824.
- Liao YY, Xu PW, Kwan KY, Ma ZY, Fang HY, Xu JY, Wang PL, Yang SY, Xie SB, Xu SQ, et al. 2019. Draft genomic and transcriptome resources for marine chelicerate Tachypleus tridentatus. Sci Data. 6(1):190029.
- Liew PL, Ng WL, Tan SG. 2015. Levels and patterns of genetic variation in an Asian horseshoe crab species, Tachypleus gigas Müller, from the Malay Peninsula. Mar Biol Res. 11:879–886.
- Lou F, Song N, Han Z, Gao T. 2020. Single-molecule real-time (SMRT) sequencing facilitates Tachypleus tridentatus genome annotation. Int J Biol Macromol. 147:89–97.
- Manca A, Mohamad F, Ahmad A, Sofa MFAM, Ismail N. 2017. Tri-spine horseshoe crab, Tachypleus tridentatus (L.) in Sabah, Malaysia: the adult body sizes and population estimate. J Asia Pac Biodivers. 10(3):355–361.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Miyata T, Usui K, Iwanaga S. 1984. The amino acid sequence of coagulogen isolated from southeast Asian horseshoe crab, Tachypleus gigas. The J Biochem. 95(6):1793–1801.
- Mohamad F, Manca A, Ahmad A, Fawwaz M, Sofa A, Alia AA, Ismail N, 2016. Width-weight and length-weight relationships of the Tri-spine Horseshoe Crab, Tachypleus tridentatus (Leach 1819) from two populations in Sabah, Malaysia: implications for population management. J Sustain Sci Manag. 11(1):1–13.
- Obst M, Faurby S, Bussarawit S, Funch P. 2012. Molecular phylogeny of extant horseshoe crabs (Xiphosura, Limulidae) indicates Paleogene diversification of Asian species. Mol Phylogenet E. 62(1):21–26.
- Pentinsaari M, Salmela H, Mutanen M, Roslin T. 2016. Molecular evolution of a widely-adopted taxonomic marker (COI) across the animal tree of life. Sci Rep. 6(1):1–12.
- Periasamy R, Ingole B, Meena RM. 2017. Phylogeny and genetic variation within population of Tachypleus gigas (Müller, 1785). Curr Sci. 112(10):2029–2033.
- Razak MRM, Kassim Z, Sabuti AA, Ismail A. 2017. Feeding ecology and food preferences of Cherok Paloh, Pahang horseshoe crab, Tachypleus gigas. Mal J Fund Appl Sci. 13(3):198–202.
- Rozihan M, Ismail E. 2012. Impact of Malaysian continental drift on the genetic diversity of horseshoe crab inferred through mtDNA sequence analysis. Int J Biol. 4(1):104–110.
- Rozihan M, Ismail E. 2011. Impact of Malaysian continental drift on the genetic diversity of horseshoe crab inferred through mtDNA sequence analysis. Int J Biol. 4(1):104.
- Sekiguchi K, Sugita H. 1980. Systematics and hybridization in the four living species of horseshoe crabs. Evolution. 34(4):712–718.
- Shingate P, Ravi V, Prasad A, Tay B-H, Garg KM, Chattopadhyay B, Yap L-M, Rheindt FE, Venkatesh B. 2020. Chromosome-level assembly of the horseshoe crab genome provides insights into its genome evolution. Nat Commun. 11(1):1–13.
- Shishikura F, Nakamura S, Takahashi K, Sekiguchi K. 1982. Horseshoe crab phylogeny based on amino acid sequences of the fibrino‐peptide‐like peptide C. J Exp Zool. 223(1):89–91.
- Vestbo S, Obst M, Quevedo FF, Intanai I, Funch P. 2018. Present and potential future distributions of Asian horseshoe crabs determine areas for conservation. Front Mar Sci. 5:164.
- Xia X. 2000. Phylogenetic relationship among horseshoe crab species: effect of substitution models on phylogenetic analyses. System Biol. 49(1):87–100.
- Zhou Y, Liang Y, Yan Q, Zhang L, Chen D, Ruan L, Kong Y, Shi H, Chen M, Chen J, et al. 2020. The draft genome of horseshoe crab Tachypleus tridentatus reveals its evolutionary scenario and well-developed innate immunity. BMC Genom. 21(1):1–15.
- Zhu G, Yuan X, Fan J. 2020. Insight into intraspecific niche divergence and conservatism in American horseshoe crabs (Limulus polyphemus). Glob Ecol Conserv. 22:e00896.
