Abstract
The mitochondrial genome of the Disckless-fingered Odorous Frog, Odorrana grahami (Anura: Ranidae), was sequenced using high-throughput sequencing technology. The genome length was 17864 bp, including 22 tRNA genes, 13 protein-coding genes, 2 rRNA genes and 1 control region (D-loop). The AT content of the mitochondrial genome was 55.9%. The composition of mitochondrial genome of O. grahami is similar to that of other species of the genus Odorrana. Phylogenetic analysis of the mitochondrial genomes of six congeners shows that O. grahami is sister to O. margaretae, but the analysis using 16S rRNA gene of additional congeners do not resolve their relationships.
Odorrana grahami (Boulenger. 1917) is part of an ancestral species group of the genus Odorrana (Anura: Ranidae; Chen et al. Citation2013), and it is diagnosed by most congeners by the lack of obvious adhesive pads at the end of the fingers (Boulenger. Citation1917). The species lives in small and medium-sized mountain streams at an altitude of 1720–3200 m, and it is mainly distributed in Sichuan, Yunnan, Guizhou, Shanxi, and Hunan Provinces of China (Fei et al. Citation2009, Citation2012; Chen et al. Citation2013). With the development of integrative taxonomic methods that utilize molecular genetic data, the taxonomy of Chinese amphibians has gone through major changes in the past decades (Wang et al. Citation2020), particularly of the genus Odorrana (Liu et al. Citation2021; Zhang et al. Citation2021), and the molecular genetic studies have revealed hidden evolutionary histories that were previously undetected (Qiao et al. Citation2018). However, the phylogenetic relationships among congeners remain unresolved in many cases (Liu et al. Citation2021), which is partly due to the lack of comparative genetic data and the limitation on the available genes. Here, we firstly reported the complete mitochondrial genome of O. grahami, which would better our understanding of the mitochondrial genome of the genus, help with the primers designs of mitochondrial genes of the genus Odorrana, and eventually facilitate the taxonomic and evolutionary researches of the group in the future.
We collected a sample of O. grahami (specimen SWFU 003918) from Daweishan National Nature Reserve, Pingbian Miao Autonomous County, Yunnan Province, China (N103°70′, E22°91′). The liver tissue was stored with 95% ethanol at −20 °C in the herbarium of Southwest Forestry University, Kunming, China (contact with Yang Wen, [email protected]). Genomic DNA of O. grahami was extracted using the DNAsecure Plant Kit (TIANGEN, Beijing, China). We used an Illumina HiSeq 2500 to perform paired-end sequencing of the sample DNA. After obtaining the sequencing data, the sequencing quality was first observed by FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and NGSQC (Dai et al. Citation2010) software was used to quality control the sequencing data according to the observed sequencing quality. Then, using the SPAdes (version 3.9.0) software with the default parameter and no cut-off parameter, we splice all the scaffolds we could put together in clean data. This software mainly constructs contig based on DBG algorithm, interrupts read into Kmers, and splices multiple Kmers. Then, Price and Mitobim were used for extended merge stitching, and the number of iterations was selected to be 50. Finally, the mitochondrial genome was annotated by MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) software (Bernt et al. Citation2013) and then submitted to GenBank (accession number MW551527).
The mitochondrial genome of O. grahami is a circular genome with a length of 17,864 bp. Including 22 tRNA genes (trnH-GTG, trnE-TTC, trnS-GCT, trnR-TCG, trnG-TCC, trnK-TTT, trnD-GTC, trnS-TGA, trnY-GTA, trnC-GCA, trnN-GTT, trnA-TGC, trnW-TCA, trnM-CAT, trnQ-TTG, trnI-GAT, trnL-TAA, trnV-TAC, trnF-GAA, trnP-TGG, trnT-TGT, and trnL-TAG), 2 rRNA genes (rrnL and rrnS), 13 protein-coding genes (PCGs) (CYTB, ND6, ND5, ND4, ND4L, ND3, COX3, ATP6, ATP8, COX2, COX1, ND2, and ND1) and 1 control region (D-loop) (). The composition of the mitochondrial genome of the O. grahami is similar to that of other species of the genus Odorrana, such as Odorrana wuchuanensis (Huang et al. Citation2016) and Odorrana graminea (Jin et al. Citation2020).
Table 1. The mitochondrial genome organization of O. grahami.
The AT content of the mitochondrial genome was 55.9%, and the base contents were: A 28.3%, C 15.5%, G 28.6%, T 27.6%, respectively. In addition to ND6, D-loop, and 8 tRNA genes (trnE-TTC, trnS-TGA, trnY-GTA, trnC-GCA, trnN-GTT, trnA-TGC, trnQ-TTG, and trnP-TGG), most of the genes in the mtDNA of O. grahami were distributed in the heavy (H) strand. Among the 13 PCGs in the mitochondrial genome, 10 genes (CYTB, ND6, ND5, ND4, ND4L, ND3, COX3
, ATP8, COX2, and ND1) have the start codon ATG, while the start codon of ATP6, COX1, and ND2 genes are ATA, GTG, and ATT, respectively. In addition, 4 of the 13 PCGs (CYTB, ND5, ND4L, and ND2) used TAG as the stop codon, 2 genes (ND6 and COX1) used AGG as the stop codon, 2 genes (ND4 and ATP8) used TAA as the stop codon, and COX2 used AGA as the stop codon. The ND3 gene was terminated by an incomplete stop codon (single stop nucleotide A), and the other 3 genes (COX3, ATP6, and ND1) were terminated by single stop nucleotide T. Among the 13 PCGs, the shortest gene was ATP8 (168 bp), and the longest gene was ND5 (1794 bp). The length of 22 tRNA genes varied from 64 to 74 bp. The lengths of rrnS, rrnl, and D-loop were 935 bp, 1582 bp, and 2220 bp, respectively. The establishment of the complete mitochondrial genome of O. grahami will provide reliable genetic data for the further study of genetic evolution, phylogeographic structure, and molecular evolution of this species.
Mitochondrial genomes of seven species of Ranidae and mitochondrial 16S rRNA genes of eight species of Odorrana were downloaded from NCBI and used for phylogenetic analyses. Rana omeimontis and Amolops wuyiensis were used as the outgroups for mitochondrial genomes phylogenetic analysis, while O. anlungensis and O. lungshengensis were used as the outgroups for 16S rRNA phylogenetic analysis. Phylogenetic relationships were reconstructed using the maximum likelihood (ML) analysis based on the above two sets of genetic data, using RAxML (Stamatakis et al. Citation2008). Genetic data were partitioned by genes, and jModelTest 0.1.1 (Darriba et al. Citation2012) was used to calculate the optimal replacement model for each partition in the two sets of sequences respectively, which was GTR + G.
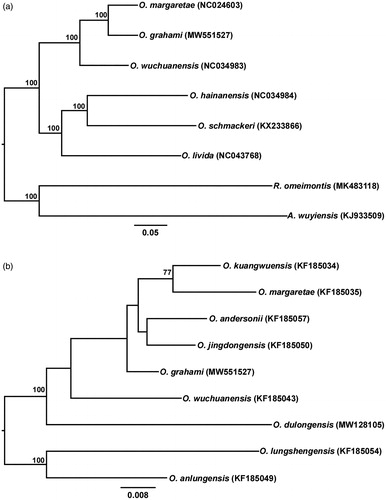
The resulting phylogenetic trees based on the mitochondrial genome suggest that O. grahami sister to O. margaretae (). Similar to previous studies (Liu et al. Citation2021), the results based on 16S rRNA gene sequences do not resolve the phylogenetic relationship of O. grahami with respect to O. kuangwuensis, O. margaretae, O. andersonii, O. jingdongensis, O. wuchuanensis, and O. dulongensis ().
Figure 1. (a) Phylogenetic relationships of six Odorrana species based on available mitochondrial genomes using ML analysis. The values above branches represent bootstrap support values. The scale bar represents 0.05 nucleotide substitutions per site. Rana omeimontis (MK483118) and Amolops wuyiensis (KJ933509) were used as outgroups. (b) Phylogenetic relationships with an expended taxa sampling among closely related species of O. grahami inferred from 16S rRNA gene tree using ML analysis. The values above branches represent bootstrap support values. The scale bar represents 0.008 nucleotide substitutions per site. Oodorrana lungshengensis (KF185054) and O. anlungensis (KF185049) were used as outgroups.
Disclosure statement
The authors report no conflicts of interest. The authors are responsible for the content and alone writing of this paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW551527.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boulenger GAL. 1917. Descriptions of new frogs of the genus Rana. J Nat Hist. 20(120):413–418.
- Chen XH, Chen Z, Jiang JP, Qiao L, Lu YQ, Zhou K, Zheng GM, Zhai XF, Liu JX. 2013. Molecular phylogeny and systematics of the genus Odorrana (Amphibia, Anura, Ranidae) inferred from two mitochondrial genes. Mol Phylogenet Evol. 69(3):1196–1202.
- Dai M, Thompson RC, Maher C, Galindo RC, Kaplan MH, Markovitz DM, Omenn G, Meng F. 2010. NGSQC: cross-platform quality analysis pipeline for deep sequencing data. BMC Genom. 11(4):S7–S9.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. JMODELTEST 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Fei L, Hu SQ, Ye CY, Huang YZ. 2009. Fauna sinica (amphibia): anura. Beijing (China): Science Press.
- Fei L, Ye CY, Jiang JP. 2012. Colored atlas of Chinese amphibians and their distributions. Chengdu (China): Sichuan Publishing House of Science and Technology.
- Huang YJ, Zhao W, Bao XK, Lin YH, Ran JC. 2016. Sequence and analysis of the complete mitochondrial genome of the Wuchuan odorous frog Odorrana wuchuanensis (Anura: Ranidae). Mitochondrial DNA Part B-Resour. 1(1):757–758.
- Jin XX, Li WY, Hu SJ, Li WM, Yang LC. 2020. The complete mitochondrial genome of large odorous frog, Odorrana graminea (Amphibia: Ranidae) and phylogenetic analysis. Mitochondrial DNA Part B-Resour. 5(3):3139–3140.
- Liu XL, He YH, Wang YF, Beukema W, Hou SB, Li YC, Che J, Yuan ZY. 2021. A new frog species of the genus Odorrana (Anura: Ranidae) from Yunnan, China. Zootaxa. 4908(2):263–275.
- Qiao L, Wen GN, Qi Y, Lu B, Hu JH, Song ZB, Fu JZ. 2018. Evolutionary melting pots and reproductive isolation: a ring‐shaped diversification of an odorous frog (Odorrana margaratea) around the Sichuan Basin. Mol Ecol. 27(23):4888–4900.
- Stamatakis AP, Hoover J., Rougemont 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57(5):758–771.
- Wang K, Ren JL, Chen HM, Lyu ZT, Guo XG, Jiang K, Chen JM, Li JT, Guo P, Wang YY, et al. 2020. The updated checklists of amphibians and reptiles of China. Biodiv Sci. 28(2):189–218.
- Zhang B, Li Y, Hu K, Li PP, Gu ZR, Xiao NW, Yang DD. 2021. A new species of Odorrana (Anura, Ranidae) from Hunan Province. ZooKeys. 1024:91–115.
