Abstract
In this study, the complete mitochondrial genome of Microconidiobolus nodosus was sequenced which is the first mitochondrial genome of the genus. The mitochondrial genome is 31,638 bp long and 27.18% in GC ratio, and it contains 14 conserved protein-coding genes, 2 ribosomal RNAs and 22 transfer RNAs. Phylogenetic analysis showed that M. nodosus was closely related to Conodiobolus sp. This study reported the whole mitochondrial genome and character of a basal fungus M .nodosus and provided a better understanding of the phylogeny of basal fungi.
The genus Neoconidiobolus B. Huang & Y. Nie Citation(2020) was established to accommodate three species producing smaller primary conidia (mostly less than 20 μm) than other Conidiobolus spp. and producing neither microspores nor capilliconidia (Nie et al. Citation2020). In the previous study, two complete mitochondrial genomes of Capillidium heterosporum (Drechsler) B. Huang & Y. Nie Citation(2020) and Conidiobolus sp. have been reported, and their phylogenetic status was also confirmed in the lineage of Entomophthoromycotina.
The ex-type Microconidiobolus nodosus (Sriniv. & Thirum.) B. Huang & Y. Nie Citation(2020) strain ATCC 16577 was obtained from American Type Culture Collection (Manassas, USA). The genomic DNA was extracted from the mycelia using the CTAB method (Watanabe et al. Citation2010). The whole genome of M. nodosus was sequenced on an Illumina HiSeq X Ten Platform (Pacific Biosciences, Nextomics Biosciences, Co., Ltd., Wuhan, China). Low-quality bases at the ends of the sequence reads were trimmed off by the quality control and the mitogenome was assembled by NOVOPlasty (Dierckxsens et al. Citation2017) by enabling the option to circularize contigs with matching ends. The mitogenome annotation was performed with MFannot tool (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) using the mitochondrial genetic code (genetic code 4) (Zhang et al. Citation2017). Transfer-RNA annotations were further predicted and confirmed by tRNAscan-SE 1.3.1 (Lowe and Eddy Citation1997).
The mitogenome of M. nodosus (GenBank accession number MW_795365) is 31,638 bp long with the GC content of 27.18%. It contains a set of 28 protein-coding genes, 2 rRNA genes (rns and rnl) and 23 tRNA genes. Among the 28 protein-coding genes, 14 protein-coding genes were core protein-coding genes: cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6 and atp6, 13 free-standing ORFs and a gene homologous to rps3. Seven introns were found in protein-encoding genes, including 1 in nad5, 2 in cob and 4 in cox1.
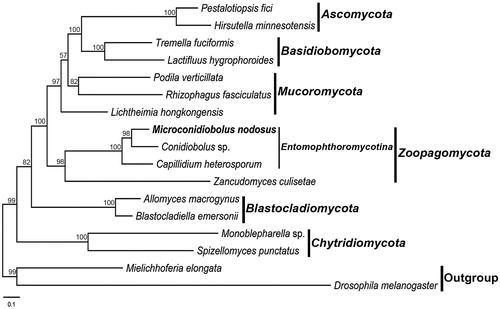
The phylogeny was inferred by the concatenated amino acid sequences of 14 proteins encoded by the conserved mitochondrial genes. The accession numbers of species used for evolutionary analysis were: Allomyces macrogynus (NC_001715), Blastocladiella emersonii (NC_011360), Capillidium heterosporum (NC_040967), Conidiobolus sp. (MN_640580), Hirsutella minnesotensis (NC_027660), Lactifluus hygrophoroides (NC_038206), Lichtheimia hongkongensis (NC_024200), Monoblepharella sp. (NC_004624), Pestalotiopsis fici (NC_031828), Podila verticillata (NC_006838), Rhizophagus fasciculatus (NC_029185), Spizellomyces punctatus (NC_003052), Tremella fuciformis (NC_036422), Zancudomyces culisetae (NC_006837). Drosophila melanogaster (NC_024511) and Mielichhoferia elongata (NC_036945). The sequences of 14 proteins were locally aligned with BioEdit (Hall Citation1999) and concatenated with SequenceMatrix (Vaidya et al. Citation2011). The final alignment length was 5,494 nucleotides and the model GTR + G + I was determined to be the most suitable evolutionary model (Li et al. Citation2021). The phylogeny was reconstructed using maximum likelihood in RAxML 8.1.17 (Stamatakis Citation2014). Microconidiobolus nodosus was grouped with Capillidium heterosporum and Conidiobolus sp. as the representative of Entomophthoromycotina in a well-supported clade (). This result was congruent with the previous studies (Nie et al. Citation2019; Sun et al. Citation2020).
Figure 1. Phylogenetic tree constructed by maximum likelihood analyses based on 14 translated mitochondrial proteins. They included oxidase subunits (Cox1, 2, and 3), the apocytochrome b (Cob), ATP synthase subunits (Atp6, Atp8, and Atp9), NADH dehydrogenase subunits (Nad1, 2, 3, 4, 5, 6, and Nad4L). The 14 fungal mitogenomes were used in this phylogenetic analysis. Drosophila melanogaster and Mielichhoferia elongata were served as outgroups. Maximum likelihood bootstrap values (500 replicates) of each clade are indicated along branches. Scale bar indicates substitutions per site.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov/). The accession number is MW_795365.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):1–9.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Li Q, Wu P, Li LJ, Feng HY, Tu WY, Bao ZJ, Xiong C, Gui MY, Huang WL. 2021. The first eleven mitochondrial genomes from the ectomycorrhizal fungal genus (Boletus) reveal intron loss and gene rearrangement. Int J Biol Macromol. 172:560–572.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Nie Y, Wang L, Cai Y, Tao W, Zhang YJ, Huang B. 2019. Mitochondrial genome of the entomophthoroid fungus Conidiobolus heterosporus provides insights into evolution of basal fungi. Appl Microbiol Biotechnol. 103(3):1379–1391.
- Nie Y, Yu DS, Wang CF, Liu XY, Huang B. 2020. A taxonomic revision of the genus Conidiobolus (Ancylistaceae, Entomophthorales): four clades including three new genera. Mycokeys. 66:55–81.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysisand post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun XR, Su D, Gui WJ, Luo F, Chen Y. 2020. Characterization and phylogenetic analysis of the complete mitochondrial genome of Conidiobolus sp. (Entomophthorales: Ancylistaceae). Mitochondrial DNA Part B. 5(1):121–122.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Watanabe M, Lee K, Goto K, Kumagai S, Sugita-Konishi Y, Hara-Kudo Y. 2010. Rapid and effective DNA extraction method with bead grinding for a large amount of fungal DNA. J Food Prot. 73(6):1077–1084.
- Zhang YJ, Zhang HY, Liu XZ, Zhang S. 2017. Mitochondrial genome of the nematode endoparasitic fungus Hirsutella vermicola reveals a high level of synteny in the family Ophiocordycipitaceae. Appl Microbiol Biotechnol. 101:3295–3244.
