Abstract
In this study, the complete chloroplast (cp) genome of Narcissus tazetta var. chinensis cv ‘Yulinglong’ was sequenced and assembled by next-generation sequencing. The complete cp genome is 159,376 in length and contained 137 genes, consisting of 91 protein-coding genes, eight ribosomal RNA genes, and 38 transfer RNA genes. Phylogenetic analyses based on chloroplast genomes highly supported that ‘Yulinglong’ was evolutionarily close to Narcissus tazetta subsp.chinensis, which may provide more desirable information for the phylogenetic relationship between Narcissus tazetta var. chinensis and its relative species.
Narcissus tazetta var. chinensis is the only variant of Narcissus L., belonging to the Amaryllidaceae (Chen et al. Citation2013). The genus originated from Late Oligocene to early Miocene, about 29.3–18.1 Ma (Santos-Gally et al. Citation2012). Chinese narcissus (Narcissus tazetta var. chinensis) is one of the ten traditional flowers in China and has high ornamental and economic values (Morikawa et al. Citation2016). It is widely distributed Fujian, Zhejiang and other southeast coastal areas in China. At present, there is the major cultivated species of Chinese narcissus called ‘Jinzhanyintai’ with six petal-like tepals surmounted by a cup-shaped corona. Recently, a variant of Chinese Narcissus named ‘Yulinglong’ with double-petaled appeared in Fujian Province of China (Ren et al. Citation2017). Cytogenetics, hybridization (Marques et al. Citation2017), and the evolution of polymorphic sexual systems (Zonneveld Citation2008) of Narcissus tazetta were widely studied. But, a detailed phylogenetic framework is still lacking. Here, the complete chloroplast (cp) genome of ‘Yulinglong’ was assembled, and it could facilitate to the study of the phylogeny, and breeding of Narcissus tazetta.
The plant material of fresh leaves tissue were obtained from Caiban Village, Jiuhu Town, Zhangzhou (Fujian, China, 117°59′56″E, 24°48′40″N), and were stored in Fujian Agriculture and Forestry University (No. FAFUYSJ01). Total genomic DNA was extracted using the optimized CTAB method (Li et al. Citation2013) and used to construct to build an Illumina pair-end library. The library was performed on an Illumina Hiseq 2000 platform (Illumina, San Diego, CA, USA) at Beijing Genomics Institute (BGI, Shenzhen, China) and generating approximately 3.4 G of raw data. Illumina sequencing data was filtered using the FastQC program (Andrews Citation2014). High-quality clean reads of around 2.6 G were applied for genome assembly by using SPAdes v 3.9.0 (http://bioinf.spbau.ru/spades) (Bankevich et al. Citation2012) and the cp genome annotation was used with the online program GeSeq (Tillich et al. Citation2017). The genome sequence of ‘Yulinglong’ has been deposited in GenBank with accession number MW322827.
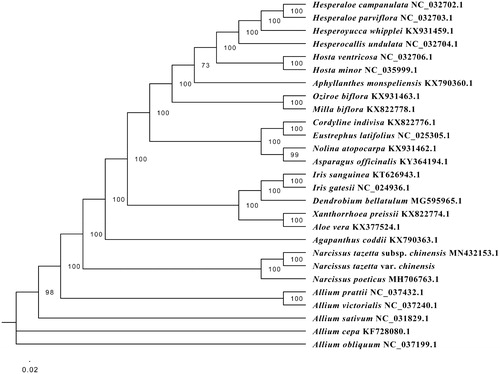
The complete circular cp genome of ‘Yulinglong’ is 159,376 bp in length, with overall GC content of 37.6%, which comprises a large single-copy region of 85,941bp, a small single-copy region of 16,451bp, and a pair of inverted repeat regions of 28,492bp. A total of 137 genes were predicted, including 91 protein-coding genes, 38 transfer RNA genes (tRNA), and 8 ribosomal RNA genes (rRNA). Among all unique genes, 15 genes tRNA(trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, ndhA) contained one intron, whereas two genes (clpP, ycf3) contained two introns. The maximum likelihood (ML) analysis was performed using RaxML software v 8.2.9, of which the bootstrap values were calculated using 1000 replicates under the GTRGAMMAI substitution model in CIPRES (Stamatakis Citation2014). The complete chloroplast sequence of 26 species was illustrated as the phylogenetic dendrogram (). The result showed that ‘Yulinglong’ is clustered with Narcissus tazetta subsp. Chinensis, which supplies available reference for Narcissus tazetta subsp. Chinensis hybridizing.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW322827.
The ‘Yulinglong’ (Narcissus tazetta var. chinensis) raw data can be found on the website (https://dataview.ncbi.nlm.nih.gov/object/SRR14141432).
Additional information
Funding
References
- Andrews S. 2014. FastQC: a quality control tool for high throughput sequence data. http://wwwbioinformaticsbabrahamacuk/projects/fastqc/.
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov A, Lesin V, Nikolenko S, Pham S, Prjibelski A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Chen HC, Chi HS, Lin LY. 2013. Headspace solid-phase microextraction analysis of volatile components in Narcissus tazetta var. chinensis Roem. Molecules. 18(11):13723–13734.
- Li XF, Jia LY, Xu J, Deng XJ, Wang Y, Zhang W, Zhang XP, Fang Q, Zhang DM, Sun Y, et al. 2013. FT-like NFT1 gene may play a role in flower transition induced by heat accumulation in Narcissus tazetta var. chinensis. Plant Cell Physiol. 54(2):270–281.
- Marques I, Fuertes Aguilar J, Martins-Louçao MA, Moharrek F, Nieto Feliner G. 2017. A three-genome five-gene comprehensive phylogeny of the bulbous genus Narcissus (Amaryllidaceae) challenges current classifications and reveals multiple hybridization events. Taxon. 66(4):832–854.
- Morikawa T, Ninomiya K, Kuramoto H, Kamei I, Yoshikawa M, Muraoka O. 2016. Phenylethanoid and phenylpropanoid glycosides with melanogenesis inhibitory activity from the flowers of Narcissus tazetta var. chinensis. J Nat Med. 70(1):89–101.
- Ren Y, Yang J, Lu B, Jiang Y, Chen H, Hong Y, Wu B, Miao Y. 2017. Structure of pigment metabolic pathways and their contributions to white tepal color formation of Chinese Narcissus tazetta var. chinensis cv Jinzhanyintai. IJMS. 18(9):1923.
- Santos-Gally R, Vargas P, Arroyo J. 2012. Insights into Neogene Mediterranean biogeography based on phylogenetic relationships of mountain and lowland lineages of Narcissus (Amaryllidaceae). J Biogeography. 39(4):782–798.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones E, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Research. 45(W1):W6–W11.
- Zonneveld BJM. 2008. The systematic value of nuclear DNA content for all species of Narcissus L. (Amaryllidaceae). Plant Syst Evol. 275(1-2):109–132.