Abstract
The complete chloroplast (cp) genome of Ehrharta erecta was sequenced and assembled for the first time. In this study, The total genome size is 134,511 bp in length and demonstrates a typical quadripartite structure containing a large single copy (LSC, 95,227 bp) and a small single copy (SSC, 12,306 bp), separated by a pair of inverted repeats (IRa, IRb) of 13,489 bp. The G + C content of this chloroplast genome was 38.76%. Gene annotation analysis identified 130 genes including 84 protein-coding genes, 38 transfer RNA, and 8 ribosomal RNA genes. The maximum-likelihood phylogenetic analysis result showed that E. erecta was closely related to O. sativa in the phylogenetic relationship.
Ehrharta erecta is a perennial grass of the grass family Poaceae native to South Africa and is a common invasive species in several places around the world, including Hawaii, Australia, New Zealand, the Mediterranean, and China (Frey Citation2005; Calvo and Moreira-Muñoz Citation2018). They are annual or perennial plants and can tolerate a wide range of abiotic conditions, such as drought conditions and high shade (Manea et al. Citation2016). E. erecta can reach heights of 60 cm, has 5- to 15-cm long leaves, and 6- to 20-cm-long panicle-like inflorescences with sessile to subsessile spikelets that look like beads on a necklace (Holloran et al. Citation2004; Grove Citation2012). In China, E. erecta are grown in large numbers and used as high-quality pasture for animals. It is important to study the chloroplast of E. erecta to increase yield.
Fresh leaves of E. erecta were collected from Panlong District, Kunming City, Yunnan Province, China (24°23′N, 102°10′E), and the voucher specimen and DNA were deposited at Qingdao University of Science and Technology (specimen code HY0812). Total genomic DNA was extracted from fresh leaves using modified CTAB (Allen et al. Citation2006), the high-quality DNA was sent to construct a genomic library and sequenced using the Illumina HiSeq platform in Novogene (Nanjing, China). About 1.2 Gb high quality, 2 × 150 bp pair-end reads were obtained and were used to assemble the complete chloroplast genome of E. erecta (Wang et al. Citation2018). The chloroplast genome of E. pilosa (Genbank accession no. MN268502) was used as a reference to assemble the complete chloroplast genome of E. erecta (Genbank accession no. MW013317) by NOVOPlasty4.2 (Dierckxsens et al. Citation2017). We also deposited the raw sequencing reads in SRA with Accession No. SRR13805805. Gene annotation was performed with the GeSeq (Michael et al. Citation2017) and manually corrected for codons and gene boundaries using the Sequin.
The complete chloroplast genome reported here is 134,511 bp in length and exhibits a typical quadripartite structure in, consisting of a pair of inverted repeat regions (IRa and IRb) with same length (13,489 bp) separated by the large single copy (LSC, 95,227 bp) and small single copy (SSC, 12,306 bp) regions. The overall GC content is 38.76%, and the corresponding values of the LSC, SSC and IR regions are 35.91, 32.31, and 43.41%, respectively. The chloroplast genome of E. erecta comprised 130 genes, including 84 protein-coding genes, 38 transfer RNA, and 8 ribosome RNA. Noticeably, nine protein-coding genes (ndhA, rpl2, rpl16, petD, petB, rpoC1, atpF, rps16, and ndhB) were disrupted by one intron, and three genes (clpP, rps12, and ycf3) by two.
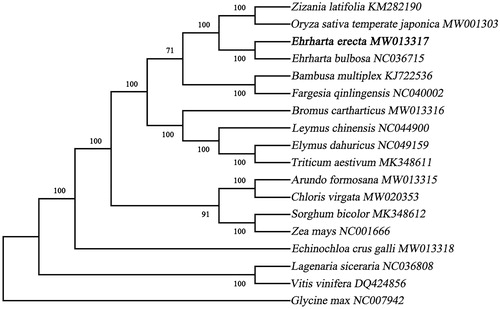
An alignment comprising the complete chloroplast genome sequences of E. erecta and other 14 related taxa of Poaceae was performed in MAFFT version 7.407 (Nakamura et al. Citation2018; Yupeng et al. Citation2020). Mosel selected process in Mega version X (Kumar et al. Citation2018) and GTR + G + I was selected by the Akaike Information Criterion. Phylogenetic tree was constructed using maximum-likelihood (ML) method and bootstrap with 1000 times iteration using the Mega version X (). The phylogenetic analysis results clearly showed that E. erecta was belonged to Poaceae and closer to O. sativa, these findings further enriched the phylogenetic relationship of the family Poaceae and will provide useful genetic information for promoting the evolutionary studies of Poaceae species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW013317. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA705412, SRR13805805, and SAMN18087868, respectively.
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethy lammonium bromide. Nat Protoc. 1(5):2320–2325.
- Calvo J, Moreira-Muñoz A. 2018. First record of Ehrharta longiflora Sm. (Poaceae, Ehrharteae) for South America. CheckList. 14(2):475–478.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty:de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Frey M. 2005. Element stewardship abstract for Ehrharta spp. Thunb. (Including Ehrharta erecta Lam, Ehrharta calycina Sm, and Ehrharta longiflora Sm). Arlington (VA): Nature Conservancy; p. 14.
- Grove S, Haubensak KA, Parker IM. 2012. Direct and indirect effects of allelopathy in the soil legacy of an exotic plant invasion. Plant Ecol. 213(12):1869–1882.
- Holloran P, Mackenzie A, Ferrell S, Johnson D. 2004. The weed workers’ handbook: a guide to techniques for removing bay area invasive plants. Richmond (CA): Watershed Project and California Invasive Plant Council; p. 29–91.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Manea A, Sloane DR, Leishman MR. 2016. Reductions in native grass biomass associated with drought facilitates the invasion of an exotic grass into a model grassland system. Oecologia. 181:175–183.
- Michael T, Pascal L, Tommaso P, et al. 2017. Geseq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(1):6–11.
- Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34(14):2490–2492.
- Wang X, Cheng F, Rohlsen D, et al. 2018. Organellar genome assembly methods and comparative analysis of horticultural plants. Hortic Res. 5(1):3.
- Yupeng W, Jiyuan L, Zhengqi F, Dongyang W, Hengfu Y, Xinlei L. 2020. Characterization of the complete chloroplast genome of Camellia brevistyla, an oil-rich and evergreen shrub. Mitochondrial DNA Part B. 5(1):386–387.