Abstract
Rhododendron datiandingense is newly reported and endemic to China. The genome of R. datiandingense is 207,311 bp in length, including a large single-copy region of 190,689 bp and a small single-copy region of 2582 bp, a pair of inverted repeat regions (IRA) of 7020 bp each. The genome encodes 110 genes, comprising 77 protein-coding genes, four ribosomal RNA genes, and 29 transfer RNA genes. Repeat analysis revealed 62 simple sequence repeats (SSRs) in the genome. Phylogenetic analysis revealed that R. datiandingense is clearly separated from the other Rhododendron species and shown in the basal position.
Rhododendron is the largest and extremely diverse genus in the heath family (Ericaceae) (Dai et al. Citation2020). It includes about 500 species in China, and more than 400 of them are endemic to China (Yang et al. Citation2020). It is naturally distributed in the North Temperate Zone, preferring moist acid soil, and northeastern Asia is its ancestral area (Yang et al. Citation2020). Rhododendron species are notable for their attractive flowers and handsome foliage and is available for garden use, but it is also used in beverages, such as azalea and Labrador tea. Rhododendron datiandingense Z. J. Feng is a newly reported species (Feng et al. Citation1996). Unlike some widely distributed congeneric species, R. datiandingense has a restricted distribution in southern China and is only found in the Guangdong Yunkaishan National Natural Reserve. Field investigation indicated that less than a hundred of individuals were found. It is considered a vulnerable species, and therefore, we report its complete chloroplast genome to provide a germplasm resource for better future conservation.
Fresh leaves of R. datiandingense were collected from the Guangdong Yunkaishan National Natural Reserve, Maoming City, China (22°17′50″N, 111°12′30″E). A voucher specimen was deposited at the Herbarium of South China Botanical Garden (Fei-Yan Zeng, [email protected]) with no. IBSC 0858371. The genomic DNA of R. datiandingense was extracted by the CTAB (cetyltrimethylammonium bromide) method, and the extracted DNA was used to construct one short-read sequencing libraries. The libraries were sequenced using the Illumina HiSeq X Ten system. The chloroplast genome of R. datiandingense was assembled by Fast-Plast 1.2.8 (McKain and Wilson Citation2017). The assembled genome was then polished by Pilon 1.24 (Walker et al. Citation2014) and annotated with CPGAVAS2 (Shi et al. Citation2019), GeSeq (Tillich et al. Citation2017), and PGA (Qu et al. Citation2019). The genome and its annotated genes were submitted to GenBank under the accession number MW381788. Phylogenetic analysis for R. datiandingense and the other 16 species was carried out with PhyloSuite 1.2.2 (Zhang et al. Citation2020) using the concatenated protein sequences of 77 chloroplast coding genes in their chloroplast genomes. PhyloSuite is an integrated phylogenetic analysis platform. Our performance, which used it, included sequence alignment with MAFFT 7.313 (Katoh et al. Citation2002), alignment trimming with trimAl 1.2 (Capella-Gutiérrez et al. Citation2009), model selection with ModelFinder (Kalyaanamoorthy et al. Citation2017), and maximum-likelihood phylogenetic inference with MrBayes 3.2.6 (Ronquist et al. Citation2012) using Nymphaea thermarum and Oryza sativa as outgroup species (Liu, Fu, et al. Citation2020).
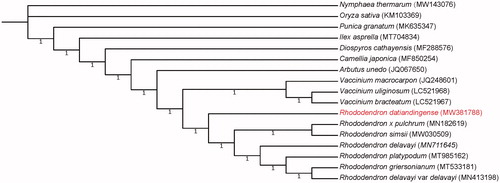
The assembled chloroplast genome of R. datiandingense was 207,311 bp in length. Its GC content was 36.06%. The genome consisted of a large single-copy region of 190,689 bp and a small single-copy region of 2582 bp, two inverted repeat regions (IRA) with 7020 bp each. The chloroplast genome encoded a total of 110 genes, including 77 protein-coding genes, four ribosomal RNA genes, and 29 transfer RNA genes. A total of 62 simple sequence repeats (SSRs) were identified in the R. datiandingense chloroplast genome (Table S1). Among them, the number of mononucleotide-repeat SSRs was 57, dinucleotide-repeat SSRs was 3, and trinucleotide-repeat SSRs was 2. Phylogenetic analysis revealed that genus Rhododendron was sister to genus Vaccinium and they were both highly supported as monophyletic, which agreed with previous phylogenetic studies (Shen et al. Citation2019; Liu, Fu, et al. Citation2020; Liu, Chen, et al. Citation2020; Ma et al. Citation2021). Within the genus Rhododendron, R. datiandingense was clearly separated from the other six Rhododendron species and shown in the basal position ().
Figure 1. Phylogenetic relationship for Rhododendron datiandingense and 16 additional species using their complete chloroplast genomes. The genomes of additional species were downloaded from GenBank and their GenBank accession numbers are shown in parentheses. The numbers on the branches are Bayesian posterior probabilities.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome sequences of Rhododendron datiandingense have been deposited in GenBank under the accession number MW381788 and is also accessible at https://doi.org/10.13140/RG.2.2.25447.78243. The associated BioProject, SRA, and Bio-Sample numbers for short reads are PRJNA686449, SRR13808853, and SAMN17118256.
Additional information
Funding
References
- Capella-Gutiérrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25:1972–1973.
- Dai X-Y, Yang C-H, Yang B, Chen P, Ma Y-P. 2020. A new species of Rhododendron (Ericaceae) from Guizhou. PhytoKeys. 146:53–59.
- Feng Z, Wu Z, Li Z. 1996. New species of Ericaceae from Guangdong. J South China Agric Univ. 17(1):59–60.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Liu J, Chen T, Zhang Y, Li Y, Gong J, Yi Y. 2020. The complete chloroplast genome of Rhododendron delavayi (Ericaceae). Mitochondrial DNA B Resour. 5(1):37–38.
- Liu D, Fu C, Yin L, Ma Y. 2020. Complete plastid genome of Rhododendron griersonianum, a critically endangered plant with extremely small populations (PSESP) from southwest China. Mitochondrial DNA B Resour. 5(3):3086–3197.
- Ma L-H, Zhu H-X, Wang C-Y, Li M-Y, Wang H-Y. 2021. The complete chloroplast genome of Rhododendron platypodum (Ericaceae): an endemic and endangered species from China. Mitochondrial DNA B Resour. 6(1):196–197.
- McKain M, Wilson M. 2017. Fast-Plast: rapid de novo assembly and finishing for whole chloroplast genomes. https://github.com/mrmckain/Fast-Plast.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542..
- Shen JS, Li XQ, Zhu XT, Huang XL, Jin SH. 2019. Complete chloroplast genome of Rhododendron pulchrum, an ornamental medicinal and food tree. Mitochondrial DNA B Resour. 4(2):3527–3528.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963.
- Yang B, Zhang G, Guo F, Wang M, Wang H, Xiao H. 2020. A genomewide scan for genetic structure and demographic history of two closely related species. Front Plant Sci. 11:1093.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
