Abstract
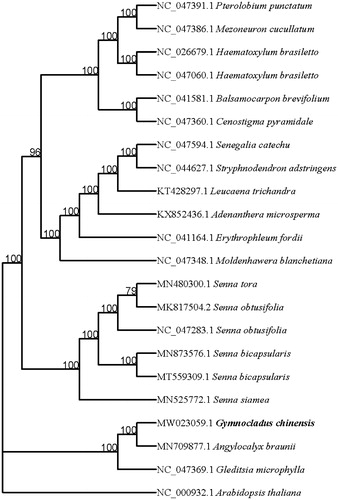
Gymnocladus chinensis Baill (Fam.: Leguminosae; Trib.: Caesalpinieae) are widely distributed in China. In this study, we assembled the complete chloroplast (cp) genome of G. chinensis. The total cp genome size was 165,315 bp in length, containing a large single-copy region of 92,356 bp, a small single-copy region of 20,449bp, and a pair of inverted repeat regions of 26,255 bp. The all GC content of G. chinensis cp was 34.95%. It encodes a total of 105 unique genes, including 75 protein-coding genes, 26 tRNA genes, and four rRNA genes. Seventeen genes contain a single intron, and two genes (ycf3 and clpP) have two introns. Phylogenetic analysis results strongly supported that G. chinensis was closely related to Angylocalyx braunii.
Gymnocladus chinensis Baill (Fam.: Leguminosae; Trib.: Caesalpinieae) is a deciduous tree, thornless, no thorns, hybrid flowers, leaves pinnae opposite, subopposite, or alternate, 5–10 pairs. Its seed oil can be used as industrial oil such as paint. Gymnocladus chinensis is an excellent tree species for ecological environment forests, economic forests, urban landscape forests, and rural ‘four sides’ greening (Yuan et al. Citation2011; Wu et al. Citation1994–2013).
The chloroplast (cp) genome has a conserved DNA of closed-loop structure, and multiple copies exist in the cell. The seeds of G. chinensis were collected in Duyun City, Guizhou Province, China (E:107.879199; N:26.247993), and were germinated and cultivated in the laboratory. The EasyPure® Plant Genomic DNA Kit (TransGen Biotech, Beijing, China) was used to extract total genomic DNA from new leaves. The total DNA was used to generate a DNA library with an average insert size of 400 bp. The library was sequenced using the Illumina NovaSeq platform. The seed specimen was deposited at the Institute for Forest Resources & Environment of Guizhou (http://frerc.gzu.edu.cn/) under the voucher number Gc-001-1. The genome sequence of G. chinensis was assembled by GetOrganelle (Jin et al. Citation2020) and was annotated by CPGAVAS2 (Shi et al. Citation2019). The annotated cp genome sequence and raw reads have been deposited in Genbank with accession number MW023059.1 and SRR12959829, respectively.
The G. chinensis cp genome showed a typical circular double-stranded tetrad structure, which was 165,315 bp, containing a large single-copy (LSC) region of 92,356 bp, a small single-copy (SSC) region of 20,449 bp, two reverse repeated regions (IRa and IRb) of 26,255 bp in length. The GC content of G. chinensis cp was 34.95%. It encodes a total of 105 unique genes, including 75 protein-coding genes, 26 tRNA genes, and four rRNA genes. Seventeen genes (trnK-UUU, rps16, trnT-CGU, atpF…) contain a single intron, and two genes (ycf3 and clpP) have two introns.
Twenty-one complete cp genome sequences including G. chinensis were aligned by MAFFT (Katoh and Standley Citation2013). Phylogenetic analysis was conducted based on maximum likelihood (ML) analyses implemented in iqtree (Nguyen et al. Citation2015) under the TVM + F+R3 nucleotide substitution model with 10,000 bootstraps. Phylogenetic relationships analysis suggested that G. chinensis is closely clustered with Angylocalyx braunii (Fam.: Leguminosae; Trib.: Papilionoideae) (). This study provided basic data for the chloroplast genetic resources and population evolution of G. chinensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. MW023059.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA672778, SRR12959829, and SAMN16576583 respectively.
Additional information
Funding
Reference
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Wu Z, Raven PH, Hong D (eds.) 1994–2013. Flora of China. 25 vols. Science Press, Beijing & Missouri Botanical Garden Press, St. Louis. Online at http://www.floraofchina.org.
- Yuan WS, Duan FZ, Wang H. 2011. [Cultivation technique of Gymnocladus chinensis]. Anhui Forestry Sci Technol. 37(6):65–66. Chinese.