Abstract
Heteropolygonatum ginfushanicum is an endemic epiphytic herb in China. The complete chloroplast (cp) genome of H. ginfushanicum is reported in this study. The total length of the cp genome is 155,508 bp with a typical quadripartite structure consisting of a large single copy region (LSC) of 84,552 bp and a small single copy region (SSC) of 18,528 bp, separated by a pair of 26,214 bp inverted repeats (IRs). It encodes a total of 113 unique genes, including 79 protein-coding, 30 tRNA, and four rRNA genes. Phylogenetic analysis indicated that H. ginfushanicum is sister to Heteropolygonatum marmoratum within subfamily Nolinoideae.
The genus Heteropolygonatum M. N. Tamura & Ogisu is a member of tribe Polygonateae of subfamily Nolinoideae in Asparagaceae (Tamura et al. Citation1997; Seberg et al. Citation2012). Species of Heteropolygonatum had been placed in the genus Polygonatum Mill. (Chao et al. Citation2013; Floden Citation2014a,b). Based on imbricate petals and the basic chromosome number of x = 16, Tamura et al. (Citation1997) separated Heteropolygonatum from Polygonatum. Phylogenetic analyses also support this treatment, showing a sister relationship between the two genera (Xiao et al. Citation2017; Floden and Schilling Citation2018). Heteropolygonatum includes about 12 species and is mainly distributed in China and adjacent Vietnam (Tamura et al. Citation2000; Xiao et al. Citation2017; Floden Citation2018). Although four cp genomes of the genus have been reported, the plastome of Heteropolygonatum ginfushanicum was not involved and genome features of the genus are still unclear (Floden and Schilling Citation2018). In this study, the complete cp genome of H. ginfushanicum was sequenced to provide basic plastome features of Heteropolygonatum, which will contribute to systematics and phylogenetic study of the Heteropolygonatum.
The sample of Heteropolygonatum ginfushanicum was collected from Siping (107°34'48.42″E, 29°9'1.80″N, elevation 1464 m), Yangxi, Daozhen, Zunyi, Guizhou, China. Fresh leaves were put into silica gel to preserve until DNA extraction and the voucher specimens were deposited in the herbarium of the Natural Museum of Guizhou University (Voucher: Hu et al. 654, GACP). Total genomic DNA was extracted according to a modified CTAB method (Doyle and Doyle Citation1987). Paired-end (PE) reads of 150 bp was conducted on an Illumina Hiseq-2500 platform at BGI-Wuhan. Approximately, 2 GB raw data (13,676,984 Clean Reads) was generated and deposited in Sequence Read Archive (SRA) under accession number SRR13587437. Then paired-end reads of the clean data was filtered and assembled de novo using the GetOrganelle script with a mean coverage of 146 ×(Jin et al. Citation2020). The chloroplast genome was annotated using program PGA (Qu et al. Citation2019) with Polygonatum odoratum (NC_050926) (Du et al. Citation2020) as a reference, then coupled with manual adjustment using Geneious v.10.1.3 (Kearse et al. Citation2012). Analysis of boundaries between IRs and single copy regions was performed by online program IRSCOPE (Amiryousefi et al, Citation2018). Finally, the annotated complete cp genome of H. ginfushanicum was submitted to NCBI GenBank (Accession number: MW363694) and the circular genome map was generated with online program CHLOROPLOT (Zheng et al. Citation2020).
The complete cp genome of Heteropolygonatum ginfushanicum is 155,508 bp in length, and has a common quadripartite structure with a large single copy (LSC) of 84,552 bp and a small single copy (SSC) of 18,528 bp separated by a pair of inverted repeats (IRs) of 26,214 bp. The plastome of H. ginfushanicum is predicted to contain 113 unique genes, including a set of 79 protein-coding, 30 tRNA and four rRNA genes, of which 20 genes were duplicated in the IR regions. Among them, eight protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rpl2 and rps16) and five tRNA genes (trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) contain one intron, and three genes (clpP, rps12 and ycf3) include two introns. The overall GC content is 37.60%, while the corresponding value in the LSC, SSC, and IR regions is 35.61%, 31.40%, and 42.99%, respectively (). As reported in other angiosperm (Mehmood et al. Citation2019, Citation2020a, Citationb; Su et al. Citation2019, Citation2021), the IR regions have the highest GC content due to the presence of rRNAs containing high GC content. Analysis of boundaries between the IRs and single copy regions of H. ginfushanicum find that the rps19 gene is 17 bp away from the LSC/IRb junction; The ndhF crosses the IRb/SSC boundary with a length of 29 bp in IRb and 2,182 bp in SSC and the ycf1 gene spans the SSC/IRa boundary with a length of 890 bp in SSC which results in a pseudogene (ψycf1) at the IRa/SSC border ().
Figure 1. Chloroplast genome map of Heteropolygonatum ginfushanicum. The species name and specific information regarding the genome length, GC content are depicted in the center of the plot.
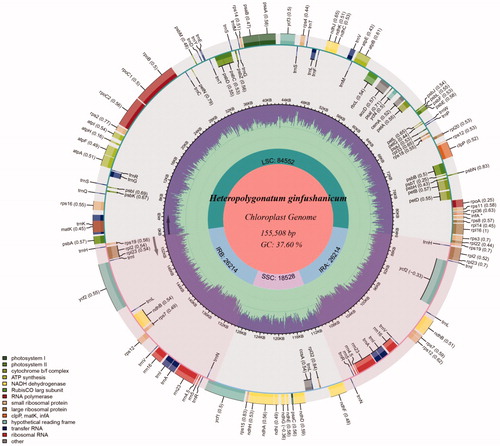
Figure 1. Chloroplast genome map of Heteropolygonatum ginfushanicum. The species name and specific information regarding the genome length, GC content are depicted in the center of the plot.
Figure 2. The boundary information between single copy and IR regions of chloroplast genomes of Heteropolygonatum ginfushanicum.
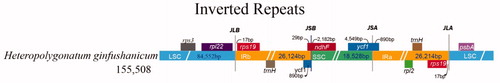
Figure 2. The boundary information between single copy and IR regions of chloroplast genomes of Heteropolygonatum ginfushanicum.
To explore the phylogenetic position of Heteropolygonatum ginfushanicum across the Asparagaceae, complete cp genomes of H. ginfushanicum and other 27 species of 7 subfamilies within Asparagaceae were selected to conduct analyses, using Lycoris aurea (MN158985) and Lycoris squamigera (NC_040164) from Amaryllidaceae as outgroups. Multiple sequence alignment of cp genome sequences were performed using MAFFT7.409 (Katoh and Standley Citation2013). Maximum likelihood (ML) analyses was conducted using RAxML-HPC2 on XSEDE v.8.2.12 (Stamatakis Citation2014) as implemented on the CIPRES Science Gateway (http://www.phylo.org/) (Miller et al. Citation2010) under the GTRGAMMA model. Bootstrap iterations (–#|–N) was set to 1000, and other parameters followed default settings.
Molecular phylogenetic analysis based on the cp genome sequences indicated that both Polygonatum and Heteropolygonatum are monophyletic and form a sister relationship within subfamily Nolinoideae, and Heteropolygonatum ginfushanicum is sister to H. marmoratum (). This finding supports the separation of Heteropolygonatum as a distinct genus from Polygonatum (Meng et al. Citation2014; Xiao et al. Citation2017; Floden and Schilling Citation2018).
Figure 3. The maximum likelihood tree of Asparagaceae inferred from 30 complete chloroplast genomes with Lycoris aurea and L. squamigera (Amaryllidaceae) as outgroups. The position of Heteropolygonatum ginfushanicum is highlighted in bold.
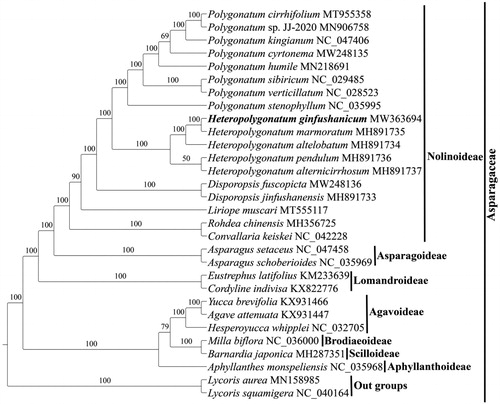
Figure 3. The maximum likelihood tree of Asparagaceae inferred from 30 complete chloroplast genomes with Lycoris aurea and L. squamigera (Amaryllidaceae) as outgroups. The position of Heteropolygonatum ginfushanicum is highlighted in bold.
Supplemental Material
Download MS Word (357.6 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data supporting this study are openly available in GenBank nucleotide database, https://www.ncbi.nlm.nih.gov/nuccore/MW363694, Associated BioProject, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA698082, BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/ SAMN17705177 and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/ SRR13587437.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Amiryousefi A, Hyvönen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34(17):3030–3031.
- Chao CT, Tseng YH, Tzeng HY. 2013. Heteropolygonatum altelobatum (Asparagaceae), comb. nova. Ann. Bot. Fen. 50(1–2):91–94.
- Doyle J, Doyle J. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19(1):11–15.
- Du Z, Qian J, Jiang Y, Duan B. 2020. The complete chloroplast genome of Polygonatum ordoratum (Mill.) Druce and its phylogenetic analysis. Mitochondrial DNA B Resour. 5(2):1601–1602.
- Floden AJ. 2014a. New names in Heteropolygonatum (Asparagaceae). Phytotaxa. 188(4):218–226.
- Floden AJ. 2014b. A new combination in Polygonatum (Asparagaceae) and the reinstatement of P. mengtzense. Ann. Bot. Fenn. 51(1-2):106–116.
- Floden AJ. 2018. Heteropolygonatum hainanense (Asparagaceae), a new species endemic to Hainan (China). Phytotaxa. 369(1):59–62.
- Floden A, Schilling EE. 2018. Using phylogenomics to reconstruct phylogenetic relationships within tribe Polygonateae (Asparagaceae), with a special focus on Polygonatum. Mol. Phylogenet. Evol. 129(2018):202–213.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Mehmood F, Abdullah, Shahzadi I, Ahmed I, Waheed MT, Mirza B. 2019. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 112(2):1522–1530.
- Mehmood F, Abdullah, Ubaid Z, Bao Y, Mirza B. 2020b. Comparative plastomics of Ashwagandha (Withania, Solanaceae) and identification of mutational hotspots for barcoding medicinal plants. Plants. 9(6):752.
- Mehmood F, Abdullah, Ubaid Z, Shahzadi I, Ahmed I, Waheed MT, Poczai P, Mirza B. 2020a. Plastid genomics of Nicotiana (Solanaceae): insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec Tobacco (Nicotiana rustica). PeerJ. 8:e9552.
- Meng Y, Nie ZL, Deng T, Wen J, Yang YP. 2014. Phylogenetics and evolution of phyllotaxy in the Solomon's seal genus Polygonatum (Asparagaceae: Polygonateae). Bot J Linn Soc. 176(4):435–451.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the gateway computing environments workshop (GCE), New Orleans (LA).
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Seberg O, Petersen G, Davis JI, Pires JC, Stevenson DW, Chase MW, Pillon Y. 2012. Phylogeny of the Asparagales based on three plastid and two mitochondrial genes. Am J Bot. 99(5):875–889.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Su T, Luo GL, Gu L, Liao HM, Hu GX. 2021. The complete plastome sequence of Tanakaea radicans (Saxifragaceae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 6(3):946–947.
- Su T, Yang JX, Lin YG, Kang N, Hu GX. 2019. Characterization of the complete chloroplast genome of Sparganium stoloniferum (Poales: Typhaceae) and phylogenetic analysis. Mitochondrial DNA B Resour. 4(1):1402–1403.
- Tamura MN, Chen SC, Turland NJ. 2000. A new combination in Heteropolygonatum (Convallariaceae, Polygonateae). Novon. 10(2):156–157.
- Tamura MN, Ogisu M, Xu J. 1997. Heteropolygonatum, a new genus of the tribe Polygonateae (Convallariaceae) from west China. Kew Bull. 52(4):949.
- Xiao JW, Meng Y, Zhang DG, He WQ, Zhang MH, Luo LY, Nie ZL, Chen G. 2017. Heteropolygonatum wugongshanensis (asparagaceae, polygonateae), a new species from Jiangxi Province of China. Phytotaxa. 328(2):189–197.
- Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A. 2020. Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet. 11:576124.
