Abstract
We determined the complete mitogenomes of two wigeons, Mareca penelope (16,603 bp) and Mareca falcata (16,597 bp) of the family Anatidae using Illumina next-generation sequencing to better understand phylogenetic relationships. Both species consisted of 37 genes (13 protein-coding genes), two ribosomal RNAs, 22 transfer RNAs, and one control region. A + T content (M. penelope 51.25%, M. falcata 51.2%) was slightly higher than G + C content (M. penelope 48.75%, M. falcata 48.8%). Phylogenetic analysis based on 13 concatenated PCG sequences indicated that M. penelope and M. falcata belong to the Mareca genus, with high bootstrap values.
Mareca is a genus of the wigeon of the family Anatidae. The Mareca was formerly placed in the Anas genus. However, molecular phylogeny studies found that the Anas genus was paraphyletic and the five Mareca species belonged to another clade from other Anas clades (Gonzalez et al. Citation2009). Complete mitochondrial genomes are a good way to understand phylogenetic relationships and improve the resolution of population genetic estimates. In this study, we analyzed the mitogenomes of two individuals of Mareca penelope Linnaeus, 1758 (Eurasian wigeon) and one individual of Mareca falcata Georgi 1775 (falcated duck) and compared them with sequences of mitogenomes of Anatinae species.
Specimens of two M. penelope and one M. falcata were collected from South Chungcheong Province (36° 47′ 04.0″ N, 126° 27′ 02″ E) on 23 January 2008 and 26 April 2007, respectively. Total DNA was extracted using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA), according to the manufacturer’s protocol. The DNA was stored in the National Institute of Biological Resources (NIBR, Hyung-Kyu Nam, [email protected]) in Incheon, South Korea, under the voucher numbers NIBRGR0000627739, NIBRGR0000627741, and NIBRGR0000627743.
Next-generation sequencing was conducted using the Illumina HiSeq platform at Genotech (Daejeon, South Korea). The mitochondrial genome was assembled de novo using CLC Assembler (ver. 6.5.4, Qiagen, Valencia, CA). Assembly errors and gaps were manually corrected through paired-end WGS read mapping using CLC Mapper (ver. 4.010.83648, Qiagen, Valencia, CA) (Kim et al. Citation2015), and the mitochondrial genome was annotated using the MITOS Webserver (Bernt et al. Citation2013). The nucleotide sequences of the mitogenomes were deposited in GenBank.
The mitogenomes are closed circular molecules of 16,603 bp (M. penelope, MW020581, MW020582) and 16,597 bp (M. falcata, MW020580). Both mitochondrial genomes were slightly AT-rich (51.25% average for M. penelope and 51.2% for M. falcata).
Both species possessed 37 genes encoding a total of 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNA genes (rRNAs), and a non-coding region (D-loop) located between tRNA-glu and tRNA-phe. Among these 37 genes, all those in the Mareca genus were distributed on the H-strand, except for the ND6 subunit gene and eight tRNA genes, encoded on the L-strand.
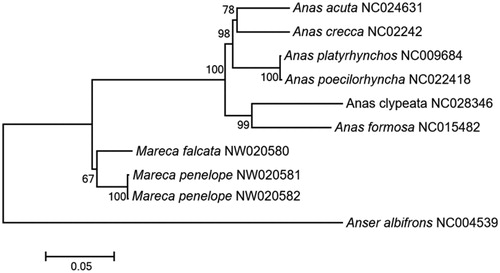
To better understand the phylogenetic relationships of mitochondrial sequences in Anatinae, we selected six Anatinae species. The best-fit partitioning scheme and nucleotide substitution model (GTR + I+G) for tree reconstruction were implemented in jModelTest v2.1.6 (Darriba et al. Citation2012). A maximum-likelihood tree was constructed using MEGA X (Kumar et al. Citation2018) with 1000 bootstrap replicates (). The phylogenetic tree indicated that the two species belonging to the genus Mareca strongly supported the monophyletic clade.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov/) under the accession no. MW020580–MW020582.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Gonzalez J, Düttmann H, Wink M. 2009. Phylogenetic relationships based on two mitochondrial genes and hybridization patterns in Anatidae. J Zool. 279(3):310–318.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5(1):1–13.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.