Abstract
We have determined the complete chloroplast genome of Chrysanthemum zawadskii Herbich isolated in Korea. The circular chloroplast genome of C. zawadskii is 151,137 bp long and has four subregions: 83,041 bp of large single copy and 18,350 bp of small single copy regions are separated by 24,873 bp of inverted repeat regions including 133 genes (87 protein-coding genes, eight rRNA genes, 37 tRNAs, and one pseudogene). There are 65 to 152 single nucleotide polymorphisms and 33 to 64 insertion and deletion regions (178 bp to 372 bp in length) identified against three available chloroplast genomes of C. zawadskii. The phylogenetic tree shows that C. zawadskii is clustered as a paraphyletic group with C. zawadskii subsp. coreanum, displaying incongruency between species and clades.
Chrysanthemum zawadskii Herbich (Asteraceae: Asteroideae), is a native plant in Korea (Park et al. Citation2020) and has economic values as traditional medicinal resources (Shin et al. Citation2010) and ornamental plants (Kim et al. Citation2014). There was controversy over its scientific name; Chrysanthemum L. and Dendranthema (DC.) Des Moul. were treated as two independent genera for a while due to morphological diversity (Bremer and Humphries Citation1993; Bremer Citation1994). However, Dendranthema was finally treated as a synonym of Chrysanthemum by a decision of the International Botanical Congress in 1999 (Trehane Citation1995; Nicolson Citation1999). Chrysanthemum including C. zawadskii has considerable variations in morphology and ploidy within species and is still unresolved based on a few chloroplast and nuclear markers (Liu et al. Citation2012). We completed C. zawadskii chloroplast genome to understand the phylogenetic position of C. zawadskii based on multiple complete chloroplast genomes.
We sequenced DNA extracted from fresh leaves of C. zawadskii (N35°34′10″, E127°1′18″; Jeongeup-si, Jeollabuk-do, Korea; InfoBoss Cyber Herbarium (IN); IB-01081; Contact: Suhyeon Park, [email protected]) using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Raw sequences obtained from Illumina NovaSeq6000 (Macrogen Inc., Korea) were filtered by Trimmomatic v0.33 (Bolger et al. Citation2014) and de novo assembled by Velvet v1.2.10 (Zerbino and Birney Citation2008). Gaps were closed with GapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17 (Li et al. Citation2009), and SAMtools v1.9 (Li Citation2013) under the Genome Information System (GeIS™; http://geis.infoboss.co.kr/). Geneious Prime® 2020.2.4 (Biomatters Ltd., Auckland, New Zealand) was used to annotate chloroplast genome based on C. indicum chloroplast (NC_020320.1; Xia et al. Citation2016).
C. zawadskii chloroplast genome (MW539687) is 151,137 bp (GC ratio is 37.5%) with four subregions: 83,041 bp of large single-copy (35.5%), 18,350 bp of small single-copy (SSC; 30.8%) regions, and 24,873 bp of a pair of inverted repeats (IR; 43.1%). It contains 133 genes (87 protein-coding genes, eight rRNAs, 37 tRNAs, and one ycf1 pseudogene in an IR region); 18 genes (seven protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in IR regions.
In comparison to Chinese C. zawadskii chloroplast (MG799556), 110 single nucleotide polymorphisms (SNPs) and 45 insertion and deletion (INDEL) regions (251 bp in length) were identified. Interestingly, 28 of 45 SNPs in 20 PCGs (62.2%) are non-synonymous SNPs (nsSNPs), similar to Chenopodium album (Park et al. Citation2021), suggesting that these variations can be used for developing molecular markers. Numbers of intraspecific variations of C. zawadskii are fewer than those identified between Korea and China samples (Heo, Kim, et al. Citation2019; Heo et al. Citation2020; Oh and Park Citation2020; Park et al. Citation2020), suggesting weak effects of geographical distribution. Additionally, numbers of intraspecific variations between our chloroplast and those of two varieties are 65 SNPs and 33 INDEL regions (178 bp in length) and 152 SNPs and 64 INDEL regions (372 bp in length), and 110 SNPs and 45 INDEL regions (251 bp in length), respectively. These variations are greater than those identified among different varieties, including Potentilla freyniana (Heo, Park, et al. Citation2019; Park, Heo, et al. Citation2019) and Aconitum barbatum (Chen et al. Citation2015), supporting that paraphyletic manner of C. zawadskii () together with larger amount of intraspecific variations than those identified from the Korean samples (Kim et al. Citation2019; Min et al. Citation2019; Park, Heo, et al. Citation2019; Park, Kim, and Xi Citation2019; Park, Kim, Xi, Oh, et al. Citation2019a, Citation2019b; Kim et al. Citation2020; Park and Oh Citation2020; Park et al. Citation2021).
Figure 1. Maximum-Likelihood phylogenetic tree and Bayesian inference tree were constructed based on sixteen Asteraceae chloroplast genomes. Phylogenetic tree was drawn based on maximum-Likelihood phylogenetic tree. Values above branches are bootstrap supports from the analysis of Maximum-Likelihood and Bayesian posterior probabilities.
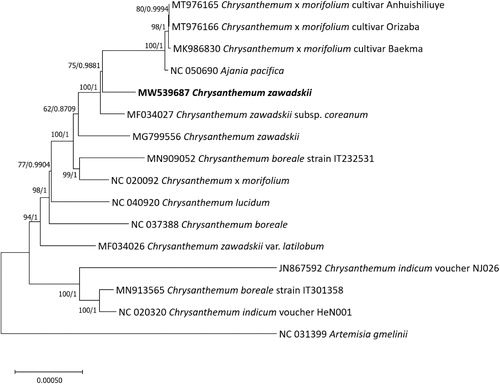
Sixteen Asteraceae complete chloroplast genomes including four C. zawadskii chloroplast genomes were aligned by MAFFT v7.450 (Katoh and Standley Citation2013) for constructing Maximum-Likelihood (ML) and Bayesian inference (BI) phylogenetic trees after adjusting SSC direction. A heuristic search was used with nearest-neighbor interchange branch swapping, the Tamura-Nei model, and uniform rates among sites to construct ML tree with 1,000 pseudo-replicates bootstrap option and default values of other options using MEGA X (Kumar et al. Citation2018). BI tree was constructed by MrBayes v3.2.6. (Ronquist et al. Citation2012). The GTR model with gamma rates was used. A Markov-chain Monte Carlo algorithm was employed for 1,100,000 generations, sampling trees every 200 generations, with four chains running simultaneously. Both trees show that two C. zawadskii chloroplast genomes are clustered with C. zawadskii subsp. coreanum and four Chrysanthemum chloroplast genomes by high supportive values (). Moreover, phylogenetic trees display three incongruencies: i) C. zawadskii (MW539687) forms a paraphyletic group with C. zawadskii subsp. coreanum and C. zawadskii (MG799556), ii) C. zawadskii var. latilobum (MF034026) is placed separately from two C. zawadskii and C. zawadskii subsp. coreanum, and iii) C. zawadskii, C. indicum, and C. boreale do not form a monophyletic group (), which is congruent to the previous phylogenetic study using chloroplast and nuclear regions (Liu et al. Citation2012). Our results present that the multiple times of evolutionary events, such as hybridization and introgression, have been occurred in Chrysanthemum genus once morphological classification is enough clear.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
Chloroplast genome sequence can be accessed via accession number of MW539687 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA688416, SAMN17175002, and SRR13320595, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Bremer K. 1994. Asteraceae: cladistics and classification. Portland (OR): Timber Press.
- Bremer K, Humphries CJ. 1993. Generic monograph of the Asteraceae-Anthemideae. Bull Nat Hist Mus Bot Ser. 23(2):71–177.
- Chen X, Li Q, Li Y, Qian J, Han J. 2015. Chloroplast genome of Aconitum barbatum var. puberulum (Ranunculaceae) derived from CCS reads using the PacBio RS platform. Front Plant Sci. 6:42.
- Heo K-I, Kim Y, Maki M, Park J. 2019. The complete chloroplast genome of mock strawberry, Duchesnea indica (Andrews) Th. Wolf (Rosoideae). Mitochondrial DNA Part B. 4(1):560–562.
- Heo K-I, Park J, Kim Y. 2019. The complete chloroplast genome of new variety candidate in Korea, Potentilla freyniana var. chejuensis (Rosoideae). Mitochondrial DNA Part B. 4(1):1354–1356.
- Heo K-I, Park J, Xi H, Min J. 2020. The complete chloroplast genome of Agrimonia pilosa Ledeb. isolated in Korea (Rosaceae): investigation of intraspecific variations on its chloroplast genomes. Mitochondrial DNA B Resour. 5(3):2264–2266.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim SJ, Lee CH, Kim J, Kim KS. 2014. Phylogenetic analysis of Korean native Chrysanthemum species based on morphological characteristics. Sci Hortic. 175:278–289.
- Kim Y, Heo K-I, Park J. 2019. The second complete chloroplast genome sequence of Pseudostellaria palibiniana (Takeda) Ohwi (Caryophyllaceae): intraspecies variations based on geographical distribution. Mitochondrial DNA Part B. 4(1):1310–1311.
- Kim Y, Park J, Chung Y. 2020. The comparison of the complete chloroplast genome of Suaeda japonica Makino presenting different external morphology (Amaranthaceae). Mitochondrial DNA Part B. 5(2):1616–1618.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997. https://arxiv.org/abs/1303.3997
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Liu P-L, Wan Q, Guo Y-P, Yang J, Rao G-Y. 2012. Phylogeny of the genus Chrysanthemum L.: evidence from single-copy nuclear gene and chloroplast DNA sequences. PloS One. 7(11):e48970.
- Min J, Park J, Kim Y, Kwon W. 2019. The complete chloroplast genome of Artemisia fukudo Makino (Asteraceae): providing insight of intraspecies variations. Mitochondrial DNA Part B. 4(1):1510–1512.
- Nicolson DH. 1999. Report of the general committee: 8. Taxon. 48(2):373–378.
- Oh S-H, Park J. 2020. The complete chloroplast genome of Euscaphis japonica (Thunb.) Kanitz (Staphyleaceae) isolated in Korea. Mitochondrial DNA B Resour. 5(3):3671–3769.
- Park J, An J-H, Kim Y, Kim D, Yang B-G, Kim T. 2020. Database of National Species List of Korea: the taxonomical systematics platform for managing scientific names of Korean native species. J Species Res. 9(3):233–246.
- Park J, Heo K-I, Kim Y, Kwon W. 2019. The complete chloroplast genome of Potentilla freyniana Bornm. (Rosaceae). Mitochondrial DNA B Resour. 4(2):2420–2421.
- Park J, Kim Y, Lee G-H, Park C-H. 2020. The complete chloroplast genome of Selaginella tamariscina (Beauv.) Spring (Selaginellaceae) isolated in Korea. Mitochondrial DNA Part B. 5(2):1654–1656.
- Park J, Kim Y, Xi H. 2019. The complete chloroplast genome sequence of male individual of Korean endemic willow, Salix koriyanagi Kimura ex Goerz (Salicaceae). Mitochondrial DNA Part B. 4(1):1619–1621.
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019a. The complete chloroplast genome of common camellia tree in Jeju island, Korea, Camellia japonica L.(Theaceae): intraspecies variations on common camellia chloroplast genomes. Mitochondrial DNA Part B. 4(1):1292–1293.
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019b. The complete chloroplast genome of common camellia tree, Camellia japonica L. (Theaceae), adapted to cold environment in Korea. Mitochondrial DNA Part B. 4(1):1038–1040.
- Park J, Min J, Kim Y, Chung Y. 2021. The comparative analyses of six complete chloroplast genomes of morphologically diverse Chenopodium album L. collected in Korea (Amaranthaceae). Int J Genomics. 2021:6643415–6643444.
- Park J, Oh S-H. 2020. A second complete chloroplast genome sequence of Fagus multinervis Nakai (Fagaceae): intraspecific variations on chloroplast genome. Mitochondrial DNA Part B. 5(2):1868–1869.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shin S, Lee Y, Moon SR, Koo IH, Hong H, Shin E, Lee M, Park J, Chung HS. 2010. Identification of secondary metabolites with antioxidant and antimicrobial activities from Artemisia iwayomogi and Chrysanthemum zawadskii. JKSABC. 53(6):716–723.
- Trehane P. 1995. Proposal to conserve Chrysanthemum L. with a conserved type (Compositae). Taxon. 44(3):439–441.
- Xia Y, Hu Z, Li X, Wang P, Zhang X, Li Q, Lu C. 2016. The complete chloroplast genome sequence of Chrysanthemum indicum. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4668–4669.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
