Abstract
Batocera lineolata Chevrolat, 1852 (Coleoptera: Cerambycidae) is an important forest pest that is found mainly in China, Vietnam, India, and Japan. The length of the complete mitochondrial genome of B. lineolate was 16,158 bp with 23.9% GC content, including 39.1% A, 14.8% C, 9.1% G and 37.0% T. The genome encoded 13 protein-coding genes, 22 tRNAs, and 2 rRNAs. Phylogenetic analysis showed that B. lineolate was closely related to Apriona swainsoni. This study provided useful genetic information for the subsequent studying the prevention of B. lineolate.
Batocera lineolata Chevrolat is an important forestry pest which usually attacks Juglans regis, Populus spp., Fraxinus spp. and Ligustrum lucidum, of which adults and larvae tunnel beneath the bark so that it can accelerate plant wilt and death (Lu and Cai Citation2016). This long horn beetle is found mainly in China, the Korean Peninsula, Vietnam, India, and Japan. While in China it has been found in Hebei, Beijing, Jiangsu, Fujian, Hunan, Sichuan and Taiwan Provinces and so on (Lu and Cai Citation2016). One generation was produced biennially or triennially with the adults overwintering (Lu and Cai Citation2016). To date, this widely distributed species has received less attention in the genetic evolution. In this study, we not only used Illumina sequencing to report the complete mitochondrial genome of B. lineolate, but also applied maximum-likelihood tree inference method to investigate the phylogenetic relationship. The results could provide an important genetic information to study the genetic evolution of B. lineolate.
The B. lineolate adults were collected from Minhou county, Fujian province, China (119.0291E, 26.1805 N) by the traps with sexual attractants. The specimens were deposited at the Fujian Agriculture and Forestry University (URL: https://lxy.fafu.edu.cn, contact person: Jiayi Ma and email: [email protected]) under the voucher number TN-202103. Total genomic DNA which extracted from an adult using TruSeq DNA sample Preparation Kit (Vazyme, China) were purified by QIAquick Gel Extraction Kit (Qiagen, GER). Nanodrop (Thermo Fisher Scientific, USA) was used to determine DNA quality and concentration. And Illumina Hiseq 2500 (Illumina, USA) was used to DNA sequencing. A total of 87,497,880 clean reads were obtained from the 89,208,078 raw reads after filtration. MitoZ and metaSPAdes (Nurk et al. Citation2017) was used to assemble the clean reads. Then using the MITOs web server (Matthias et al. Citation2013) to annotate the assembly sequence, and using tRNAscan software (Lowe and Eddy Citation1997) to predict tRNA genes. The complete mitochondrial genome sequence of B. lineolate has been submitted to NCBI GenBank with accession number MW629558. The complete mitochondria genome of B. lineolate was 16,158 bp length and encoded 13 protein-coding genes (PCGs), 22 tRNA genes, two ribosomal RNA (rRNA) genes. The GC content of the complete genome was 23.9%, including 39.1% A, 14.8% C, 9.1% G and 37.0% T.
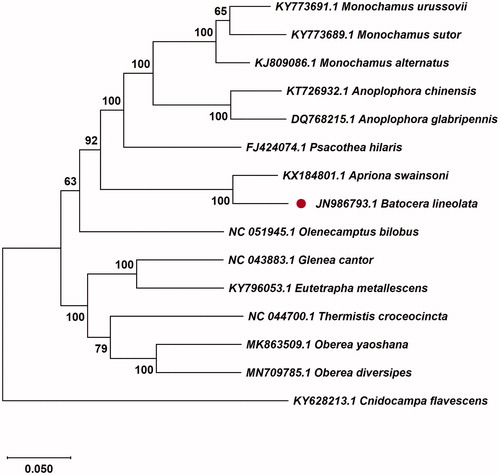
The evolutionary tree was constructed for purpose of confirming the phylogenetic position of B. lineolate with related 14 different species insects by MEGAX using Maximum Likelihood tree model with 1000 bootstrap replicates (Kumar et al. Citation2008). The B. lineolate is closely related to Apriona swainsoni (Coleoptera: Cerambycidae) through the phylogenetic tree (). The availability of the complete mitochondrial genome of B. lineolate allows further research about the genetic evolution and control tactics of B. lineolate can be facilitated.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the assession no. MW629558. The associated BioProject, SRA and Bio-Sample number was PRJNA702870, SRR13743230, and SAMN17981209, respectively.
Additional information
Funding
References
- Kumar S, Nei M, Dudley J. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9(4):299–306.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Lu JF, Cai JY. 2016. Research on biological characteristics of Batocera lineolata Chevrolat in Guizhou Province. Biol Disaster Sci. 39(02):109–112.
- Matthias B, Alexander D, Frank J, Fabian E, Catherine F, Guido F, Joern P, Martin M, Peter FS. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.