Abstract
Sapindus delavayi, widely distributed in central to southwest of China, is an economically important forest tree. The chloroplast genome is 159,861 bp in length with a typical circular quadripartite structure, containing 129 genes (84 protein-coding genes, 37 tRNAs, and eight rRNAs). Our phylogenetic result clearly showed that S. delavayi and S. mukorossi have the closest relationship.
Sapindus delavayi (Franch.) Radlk., commonly known as the soap fruit, belonging to the Sapindaceae family, is widely distributed in central to southwest of China (Sun, Jia, et al. Citation2017; Sun, Wang, et al. Citation2017). The pericarp of S. delavayi is rich in polyphenols and saponins, and the saponins can be used as a highly effective natural surfactant in commercial productions (Nakayama et al. Citation1986). The S. delavayi saponins are also used to expel parasite, suppress coughing, and treat inflammation (Quetin-Leclercq et al. Citation1992; Zhou et al. Citation2010; Zhou et al. Citation2013; Wu et al. Citation2013; Xu et al. Citation2013). It is reported that the seed oils produced by S. delavayi are rich in medium-chain monounsaturated fatty acids and suitable to produce biodiesel (Sun et al. Citation2018). The root and fruit of S. delavayi are commonly used as traditional Chinese medicine, and the trunk of the plant is used as wood to make box boards and combs (Xu et al. Citation2013). As a wild resource, S. delavayi is limited in its development and utilization due to the lack of genetic information (Zeng et al. Citation2018). To restore the S. delavayi forest and make better use of it, it is necessary to protect the existing wild S. delavayi and have a deeper understanding of its phylogenetic relationships with other Sapindaceae species.
Chloroplast genome sequences have been widely used to reconstruct phylogenetic relationships and explore the structural and functional evolution (Jansen et al. Citation2007; Moore et al. Citation2010; Wang et al. Citation2020). In our study, the complete chloroplast genome of S. delavayi was assembled and annotated for the first time. The study provides a basis for the relationship between several main clades of the Sapindaceae family.
The leaves of S. delavayi were collected from Nanjian, Yunnan, China (100°24′′08′′E, 24°54′70′′N, 1132 m), in August 2020. Total genome DNA was extracted and sequenced by Annoroad Gene Technology (Beijing, China). The original specimen of this species was stored in the herbarium of Southwest Forestry University with accession number: SWFU-YAB-H-0330. Total DNA was sequenced using the Illumina Novaseq 6000 platform at Annoroad Gene Technology (Beijing, China). About 4.51 GB of high-quality clean reads were generated by 150 bp paired read length. The complete cp genome of S. delavayi was assembled with GetOrganelle version 1.6.2 (Jin et al. Citation2020) (The parameters of GetOrganelle version 1.6.2 are as follows: Cycles: 15; KMER value: 21, 45, 65, 85, and 105; The chloroplast genome database name used for assembly: EMBPLANT_PT). Genome annotation of S. delavayi was performed by employing the program Geneious R8 and manually adjusted by comparing it to the reference genome S. mukorossi. The chloroplast DNA sequence was deposited at the GenBank database with the accession number MW041255.
The complete cp genome of S. delavayi was 159,861 bp long in length. It contained SSC of 18,657 bp add of and LSC of 85,242 bp, separated by a pair of inverted repeats (IRa and IRb, 27,981 bp), showed a typical angiosperm quadripartite structure. The GC contents of the four regions above were 31.6%, 35.8%, 42.6%, and 42.6%, respectively. The whole cp genome included functional 129 genes (eight rRNA, 37 tRNA, and 84 protein-coding genes). Three genes (rps12, ycf3, and clpP) had two introns and 17 genes (rpl2, ndhB, trnI-GAU, trnA-UGC, ndhA, trnI-GAU, ndhB, petB, trnV-UAC, trnL-UAA, rpoC1, atpF, trnG-GCC, rps16, and trnK-UUU) had only one intron.
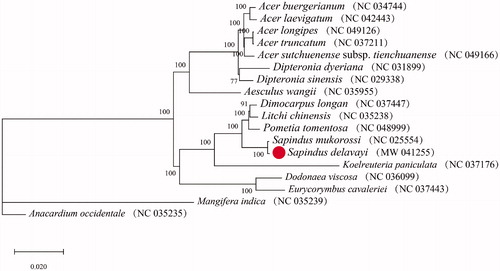
We constructed a phylogenetic tree with the cp genomes of S. delavayi and other 17 species to analyze the phylogenetic relationship of S. delavayi with species in the Sapindaceae family (the species Anacardium occidentale were set as outgroup). MEGA version 7.0 was applied to align the 18 complete chloroplast sequences (Kumar et al. Citation2016). With the Maximum-likelihood method, the model ‘K3Pu + F+R3’ was chosen to be the best nucleotide substitution model in IQ-tree version 1.5.5 (Nguyen et al. Citation2015) (). Five Acer species and Dipteronia sinensis, Dipteronia dyeriana, and Aesculus wangii formed a cluster (BS = 100). Another cluster (BS = 100) contained eight species (including the cp genome newly sequenced in this study): Dimocarpus longan, Litchi chinensis, Pometia tomentosa, Sapindus mukorossi, Sapindus delavayi, Koelreuteria paniculata, Dodonaea viscosa, Euonymus hamiltonianus. Anacardium occidentale, and Mangifera indica were using as outgroup. Within the tribe Sapindus, S. delavayi had close relationship with S. mukorossi. The analysis provided essential molecular information for S. delavayi and reference for the systematic differentiation of Sapindaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession No. MW041255. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA698468, SRR13711031, and SAMN17720602, respectively.
Additional information
Funding
References
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104(49):19369–19374.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 107(10):4623–4628.
- Nakayama K, Fujino H, Kasai R, Tanaka O, Zhou J. 1986. Saponins of pericarps of Chinese Sapindus delavayi (Pyi-shiau-tzu), a source of natural surfactants. Chem Pharm Bull (Tokyo). 34(5):2209–2213.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective Stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Quetin-Leclercq J, Elias R, Balansard G, Bassleer R, Angenot L. 1992. Cytotoxic activity of some triterpenoid saponins. Planta Med. 58(3):279–281.
- Sun CW, Jia LM, Xi BY, Wang LC, Weng XH. 2017. Natural variation in fatty acid composition of Sapindus spp. seed oils. Ind Crop Prod. 102:97–104.
- Sun CW, Wang JW, Duan J, Zhao GC, Weng XH, Jia LM. 2017. Association of fruit and seed traits of Sapindus mukorossi germplasm with environmental factors in southern China. Forests. 8(12):491.
- Sun CW, Wang LC, Liu JM, Zhao GC, Gao SL, Xi BY, Weng XH, Jia LM. 2018. Genetic structure and biogeographic divergence among Sapindus species: An inter-simple sequence repeat-based study of germplasms in China. Industrial Crops & Products, 118:1–10.
- Wang YB, Liu BB, Nie ZL, Chen HF, Chen FJ, Figlar RB, Wen J. 2020. Major clades and a revised classification of Magnolia and Magnoliaceae based on whole plastid genome sequences via genome skimming. J Syst Evol. 58(5):673–695.
- Wu H, Wang N, Weng Z, Xu DP, Wang HY, Yao WR. 2013. Researches on surface activity of Sapindus saponin product and its mixed system. Fine Chem. 30(02):149–154.
- Xu KJ, Zha-Xi CD, Ding LS. 2013. Research progress on chemical constituents and biological activities of Sapindus species. Nat Prod Res Dev. 25(02):267–273+257.
- Zeng WL, Xiong H, Ye HL, Wang LC, Dan HL, Zhou T, Duan HM, Zhao P. 2018. Selection of superior individuals of Sapindus delavayi with high content of saponins or polyphenol. J Cent South Univ Forest Technol. 37(09):86–92.
- Zhou L, Xie WS, Luo YJ. 2010. Studies on the bioactivity of the extracts from Sapindus delavayi used as a pesticide. Plant Protec. 36(5):162–164.
- Zhou L, Xie WS, Ren HT, Zhou B. 2013. Extracting process and bioactivity of saponins from Sapindus delavayi pericarp. J Yunnan Agric Univ. 28(3):433–438.