Abstract
Here, we report the complete chloroplast genome of Pittosporum brevicalyx. The genome is 153,388 bp in size, which is comprised of a large single-copy (LSC) region of 84,724 bp, a small single-copy (SSC) region of 18,716 bp, and two inverted repeat (IR) regions of 24,974 bp. The overall GC content of the plastome was 38.3%. The new sequence comprised 127 unique genes, including 82 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. Phylogenetic analysis showed that P.brevicalyx was close to Pittosporum kerrii and Pittosporum eugenioides. These data may providing useful information for phyletic evolution of P.brevicalyx within the Pittosporaceae family.
Pittosporum brevicalyx (Oliver) Gagnepain (1908) is a tree species belonging to the Pittosporaceae family, is a small evergreen shrub distributed in the Sichuan, Guangdong, Guangxi, Guizhou, Hubei, and Hunan provinces of China. The root, bark, and fruit of this shrub are used for the treatment of chronic bronchitis and coughs by local people (Feng et al. Citation2010). The crude extract of P.brevicalyx was reported to possess antituberculous activities in vitro and in vivo (Wu et al. Citation2007).
The chloroplast genome is highly conserved in plants and therefore ideal for ecological, evolutionary, and diversity studies (Wicke et al. Citation2011). To date, many studies have performed whole chloroplast genome sequencing. However, for this tree species, the classification and phylogenetic relationships of P. brevicalyx remain poorly known. In the present study, we report the complete chloroplast genome of P. brevicalyx, thus providing useful information for future studies in genetic background and evolution.
The fresh leaves were collected from Tropical and Subtropical Economic Crops Institute of Yunnan Academy of Agricultural Science, Baoshan, China. (Yunnan, China; geospatial coordinates: 99°10′25.61″E, 25°08′0.11″N; Altitude: 1655 m). The voucher specimens of P.brevicalyx were deposited at the herbarium of Tropical and Subtropical Economic Crops Institute of Yunnan Academy of Agricultural Science Baoshan, China (Contact person: Mr Mingkun Xiao, Email: [email protected]) under the voucher number YAAS-H-T-03, and DNA samples were stored at the Laboratory of Tropical and Subtropical Economic Crops Institute of Yunnan Academy of Agricultural Science Baoshan, China. The total genomic DNA was extracted by using the Magnetic beads plant genomic DNA preps Kit (TSINGKE Biological Technology, Beijing, China). We assembled the chloroplast genome using GetOrganelle v1.6.2e (Jin et al. Citation2020). Genome annotation was performed with the online annotation tool CPGAVAS2 (Shi et al. Citation2019). Finally, the chloroplast DNA sequence with complete annotation information was submitted to GenBank with accession number MW368386.
The complete genome of P.brevicalyx is 153,388 bp in size, containing a small single-copy (SSC) region of 18,716 bp and a large single-copy (LSC) region of 84,724bp separated by a pair of inverted repeat (IR) regions of 24,974 bp. The total GC content is 38.3%, while the corresponding values of the LSC, IR, and SSC regions are 36.5%, 43.4%, and 32.5%, respectively. There were 127 genes, including 82 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. Among them 13 genes(atpF, ndhA, ndhB, rpl16, rpl2, rps16, rpoC1, trnK-UUU, trnT-CGU, trnL-UAA, trnV-UAC, trnE-UUC, trnA-UGC) have single introns, while 2 genes (ycf3 and clpP) have double introns.
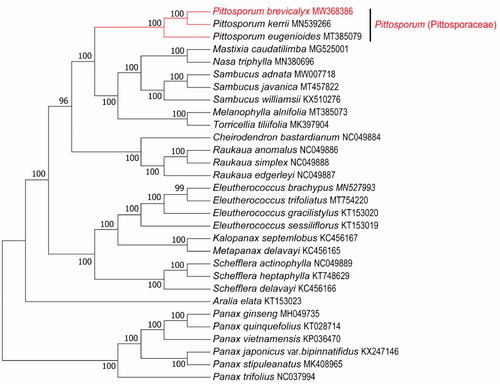
To confirm the phylogenetic relationship of P.brevicalyx in Pittosporaceae family, Phylogenetic analysis was performed based on the complete cp genomes of P.brevicalyx and other 29 Pittosporaceae species. We aligned all chloroplast genomes by using MAFFT v7.471 (Katoh et al. Citation2019) and analyzed by IQ-TREE v1.6.12 (Minh et al. Citation2020) under the TVM + F+R6 nucleotide substitution model. As illustrated in , P. brevicalyx appeared to be closely related to Pittosporum kerrii and Pittosporum eugenioides. The chloroplast genome of P.brevicalyx will provide useful genetic information for further study on genetic diversity and conservation of Pittosporaceae species.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number: MW368386. The associated Bio-Project, Bio-Sample and SRA, numbers are PRJNA688982, SAMN17192762 and SRR13349194 respectively.
Additional information
Funding
References
- Feng C, Li BG, Gao XP, Qi HY, Zhang GL. 2010. A new triterpene and an antiarrhythmic liriodendrin from Pittosporum brevicalyx. Arch Pharm Res. 33(12):1927–1932.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. https://doi.org/10.1186/s13059-020-02154-5
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von HA, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evo. 37(5):1530–1534.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Wicke S, Schneeweiss GM, Depamphilis CW, Muller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297.
- Wu GD, Qian ZQ, Dai J. 2007. Experimental study on antituberculous activities of Pittosporum brevicalyx (Oliv.) Gagnep. J Chin Antituberculosis Assoc. 29:41–43.