Abstract
We presented a complete chloroplast genome of a new species candidate of Plantago depressa, Willd. named as Plantago wonjuenesis sp. nov, which is 164,946 bp long (GC ratio is 38.0%) and has four subregions: 82,985 bp of large single copy and 4,647 bp of small single-copy regions are separated by 38,657 bp of inverted repeat regions including 94 protein-coding genes (PCGs), eight rRNAs, and 38 tRNAs. Number of variations between P. wonjuenesis and P. depressa can be considered as interspecific variations. Bootstrapped phylogenetic trees constructed with conserved 78 PCGs of eleven Plantaginaceae chloroplast genomes present that P. wonjuensis is clustered with P. depressa, P. fengdouensis, and P. media.
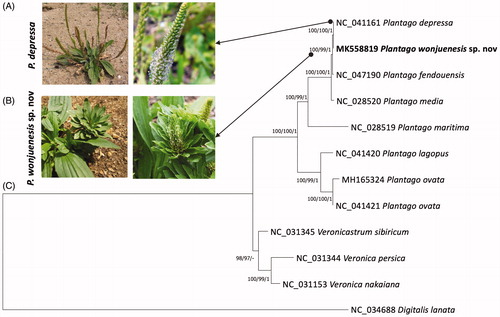
Recently, a new species candidate of Plantago depressa was identified in the Republic of Korea. It shows expended leaf-like bract unlike the small bract of P. depressa as well as short spike length and cone-shape inflorescence (). To understand the genetic difference of this new candidate species named Plantago wonjuenesis sp. nov, we completed its chloroplast genome like the previously sequenced chloroplast genomes of new species candidates (Heo et al. Citation2019; Kim et al. Citation2019; Oh et al. Citation2019; Kim et al. Citation2020) and mitochondrial genome (Park, Xi, Kim, et al. Citation2020).
Figure 1. (A,B) display the shape of P. depressa and P. wonjuenesis sp. nov, respectively. (C) presents Maximum Likelihood (bootstrap repeat is 1000), Neighbor-joining (bootstrap repeat is 10,000), and Bayesian inference (Number of generations is 1,100,000) phylogenetic trees of eleven Plantaginaceae chloroplast genomes. Phylogenetic tree was drawn based on Maximum Likelihood tree. The numbers above branches indicate bootstrap support values of Maximum Likelihood and Neighbor-joining trees and posterior probability of Bayesian inference tree.
Plantago wonjuenesis isolated in Jijeong-myeon, Wonju City, Gangwon province, Republic of Korea (37.404215 N, 127.820117E; Voucher deposited in the National Institute of Biological Resources (NIBR) Herbarium (KB); NIBRVP0000759185; Contract: Chan-Ho Park; [email protected]). Total DNA of P. wonjuenesis was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Two Gbp raw sequences obtained from Illumina HiSeq4000 at Macrogen Inc., Korea, were filtered by Trimmomatic v0.33 (Bolger et al. Citation2014) and de novo assembly and confirmation were conducted by Velvet v1.2.10 (Zerbino and Birney Citation2008), GapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17 (Li Citation2013), and SAMtools v1.9 (Li et al. Citation2009) under the environment of Genome Information System (GeIS; http://geis.infoboss.co.kr). Geneious R11 v11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used for chloroplast genome annotation based on P. depressa chloroplast genome (NC_041161; Kwon, Kim, Park, et al. Citation2019).
The chloroplast genome of P. wonjuenesis (GenBank accession is MK558819) is 164,946 bp (GC ratio is 38.0%) and has four subregions: 82,985 bp of large single copy (36.6%) and 4,647 bp of small single copy (30.2%) regions are separated by 38,657 bp of inverted repeat (IR; 39.9%) regions. It contains 140 genes (94 protein-coding genes (PCGs), eight rRNAs, and 38 tRNAs); 26 genes (15 PCGs, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions.
Based on pairwise alignment with P. depressa chloroplast genome (NC_041161; Kwon, Kim, Park, et al. Citation2019), 35 single nucleotide polymorphisms (SNPs) and 15 insertions and deletion (INDEL) regions which cover 317 bp in total are identified. Five SNPs are in one IR (0.012%), 7 SNPs in SSC (0.15%), and 18 SNPs are in LSC regions (0.022%), displaying the highest density of SNPs in SSC region. The largest INDEL region of which length is 262 bp (82.65%) is located in the border of IR and SSC regions, which majorly contributed the expansion of IR region in comparison to that of P. depressa. One 1-bp INDEL region is in SSC region and the rest 13 INDELs (54 bp; 17.03%) are in LSC region. The number of sequence variations is higher than those of species including Rosa rugosa (40 SNPs and 37 INDELs of which length is 224 bp; Kim et al. Citation2019), Potentilla freyniana (19 SNPs and 35 INDELs of which length is 198 bp; Heo et al. Citation2019), and Suaeda japonica (one SNP and two INDELs; Kim et al. Citation2020) of which morphological features are different from the original species.
Numbers of chloroplast intraspecific variations originated from samples between countries, such as Liriodendron tulipifera (12 SNPs; Park, Kim, Kwon, Xi, et al. Citation2019), Nymphaea alba (11 SNPs and 6 INDELs of which length is 23 bp; Park, Kim, Kwon, Nam, et al. Citation2019), Coffea arabica (82-bp INDEL and 2-bp INDEL; Park, Kim, Xi, et al. Citation2019), Illicium anisatum (21 SNPs and 17 INDELs of which length is 114 bp; Park, Kim, Xi Citation2019), Duchesnea chrysantha (48 SNPs and 3 INDELs covering 58 bp; Park, Kim, Lee Citation2019), Arabidopsis thaliana (10–121 SNPs and 4–46 INDELs (14–570 bp in length); Park, Xi, Kim Citation2020) and Marchantia polymorpha subsp. ruderalis (4 SNPs; Kwon, Kim, Park Citation2019), are similar to or slightly higher than those identified in this study, supporting that the number of variations can be considered as inter-species as both species are isolated in Korea.
Concatenated alignment of 78 coding genes using MAFFT v7.450 (Katoh and Standley Citation2013) from 12 Plantaginaceae chloroplast genomes including P. wonjuenesis were subjected to construct bootstrapped Neighbor-joining, Maximum Likelihood, and Bayesian inference phylogenetic trees using MEGA X (Kumar et al. Citation2018) and MrBayes v3.2.6 (Ronquist et al. Citation2012), respectively. Three phylogenetic trees confirmed that P. wonjuenesis is clustered with P. depressa, which is congruent to the previous study (Park et al. Citation2017), as well as Plantago fengdouensis and P. media supported by high supportive values (). In comparison to phylogenetic distances from P. fengdouensis, an endemic species in China (Wang et al. Citation2004), to P. wonjuenesis and P. depressa, P. wonjuenesis can be considered as a new species in both aspects of morphology and chloroplast genome.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
Chloroplast genome sequence can be accessed via accession number MK558819 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA669482, SAMN16446523, and SRR12834755, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Heo K-I, Park J, Kim Y. 2019. The complete chloroplast genome of new variety candidate in Korea, Potentilla freyniana var. chejuensis (Rosoideae). Mitochondrial DNA B. 4(1):1354–1356.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim Y, Heo K-I, Nam S, Xi H, Lee S, Park J. 2019. The complete chloroplast genome of candidate new species from Rosa rugosa in Korea (Rosaceae). ). Mitochondrial DNA B Resour. 4(2):2433–2435.
- Kim Y, Park J, Chung Y. 2020. The comparison of the complete chloroplast genome of Suaeda japonica Makino presenting different external morphology (Amaranthaceae). Mitochondrial DNA B. 5(2):1616–1618.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kwon W, Kim Y, Park J. 2019. The complete mitochondrial genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: inverted repeats on mitochondrial genome between Korean and Japanese isolates. Mitochondrial DNA Part B. 4(1):769–770.
- Kwon W, Kim Y, Park C-H, Park J. 2019. The complete chloroplast genome sequence of traditional medical herb, Plantago depressa Willd. (Plantaginaceae). Mitochondrial DNA B. 4(1):437–438.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Oh S-H, Suh HJ, Park J, Kim Y, Kim S. 2019. The complete chloroplast genome sequence of a morphotype of Goodyera schlechtendaliana (Orchidaceae) with the column appendages. Mitochondrial DNA B. 4(1):626–627.
- Park J, Kim Y, Kwon W, Nam S, Song MJ. 2019. The second complete chloroplast genome sequence of Nymphaea alba L. (Nymphaeaceae) to investigate inner-species variations. Mitochondrial DNA B. 4(1):1014–1015.
- Park J, Kim Y, Kwon W, Xi H, Kwon M. 2019. The complete chloroplast genome of tulip tree, Liriodendron tulifipera L. (Magnoliaceae): investigation of intra-species chloroplast variations. Mitochondrial DNA B Resour. 4(2):2523–2524.
- Park J, Kim Y, Lee K. 2019. The complete chloroplast genome of Korean mock strawberry, Duchesnea chrysantha (Zoll. & Moritzi) Miq. (Rosoideae). Mitochondrial DNA B. 4(1):864–865.
- Park J, Kim Y, Xi H. 2019. The complete chloroplast genome of aniseed tree, Illicium anisatum L. (Schisandraceae). Mitochondrial DNA B. 4(1):1023–1024.
- Park J, Kim Y, Xi H, Heo K-I. 2019. The complete chloroplast genome of ornamental coffee tree. Coffea arabica L. (Rubiaceae). Mitochondrial DNA B. 4(1):1059–1060.
- Park J, Park C, Kim Y. Whole genome sequence of new species of Plantago as a second whole genome in Plantaginaceae 2017. The XIX International Botanical Congress, 2017.
- Park J, Xi H, Kim Y. 2020. The complete chloroplast genome of Arabidopsis thaliana isolated in Korea (Brassicaceae): an investigation of intraspecific variations of the chloroplast genome of Korean A. thaliana. Int J Genomics. 2020:3236461.
- Park J, Xi H, Kim Y, Nam S, Heo K-I. 2020. The complete mitochondrial genome of new species candidate of Rosa rugosa (Rosaceae). Mitochondrial DNA B Resour. 5(3):3435–3455.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Wang Y, Li Z, Wu J, Huang H. 2004. Plantago fengdouensis, a new combination in the Plantaginaceae from China. Acta Phytotaxonomica Sinica. 42(6):557–560.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
