Abstract
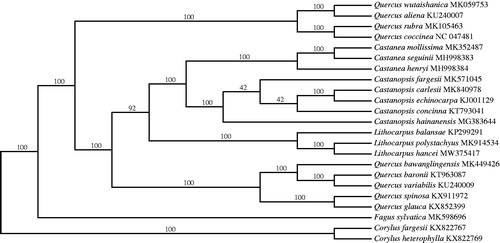
Lithocarpus hancei (Benth.) Rehd is a widely distributed evergreen tree with broad-leaves that dominates the lower stories of the forest in China. Here, we sequenced and assembled the complete chloroplast genome of L. hancei. The genome is 161,304 bp with one large single copy (LSC: 90,585 bp), one small single copy (SSC: 18,959 bp), and two inverted repeat (IR) regions (IRa and IRb, each 25,880 bp). It contains 117 genes, including 80 protein-coding genes, 33 tRNA genes, and four rRNA genes. Phylogenetic analysis of 21 representative cp genomes of the Fagaceae suggests Lithocarpus is monophyletic with strong bootstrap support and also that L. hancei is closely related to L. polystachyus. The cp genome is important for constructing a robust phylogeny of Lithocarpus and Fagaceae for future study.
Lithocarpus Blume is a large genus classified in the Fagaceae with more than 300 species that is predominantly distributed in Asia (Huang and Bruce Bartholo 1999). China hosts 122 Lithocarpus species (Huang and Bruce Bartholo Citation1999). This genus plays an essential role in the evergreen broad-leaved forest (Huang and Bruce Bartholo 1999; Cannon and Manos Citation2003). Lithocarpus hancei (Benth.) Rehd 1919 is a widely distributed evergreen tree with broad-leaves that occurs under 2600 m in central and south China (Huang and Bruce Bartholo Citation1999). It is a shade-tolerant canopy species that lives in the shaded-story or canopy gaps and sometimes is codominant the lower stories with Castanopsis Spach and Fagus L. (Cao Citation2001). Chloroplast DNA (cp DNA) has been widely used for species identification, phylogeny reconstruction, demographic history tracing and species divergence studies (Yang et al. Citation2016; Yang et al. Citation2017; Alzahrani et al. Citation2020), because of its conserved genome structure and uniparental inheritance (Birky et al. Citation1983). However, only three cp genomes of Lithocarpus are available in GenBank, of which only the cp genome of Lithocarpus balansae (Drake) A.Camus has been fully annotated. To better understand the phylogenetic position of L. hancei, in this study we sequenced and assembled its chloroplast genome.
The voucher specimen (Li Buhang et al. BSZ142) was collected from Baishanzu Mountain, Zhejiang, China (27°32'25′'N, 118°57'49′'E) and deposited at the herbarium of South China Botanical Garden (SCBG, Feiyan Zeng, [email protected]). Genomic DNA was extracted from silica gel-dried leaves using the CTAB method (Doyle Citation1987), and sequenced by the Illumina Hiseq X platform at the Beijing Genomics Institute (Wuhan, China). The clean reads were assembled using the program GetOrganelle v1.7.1 (Jin et al. Citation2020). The annotation was performed on DOGMA (Wyman et al. Citation2004) and manually corrected using Geneious 7.1.4 (Biomatters, Ltd, Auckland, New Zealand). The complete cp genome of L. hancei and 20 related species classified in the Fagaceae, plus two outgroups (Corylus heterophylla Fisch and C. fargesii Schneid) were aligned using MAFFT with auto alignment strategy (Katoh and Standley Citation2013). Phylogenetic analysis was performed using RAxML v8.2.12 with 1000 bootstrap replicates under the GTA + GAMMA model (Stamatakis Citation2014).
The complete cp genome of L. hancei is 161,304 bp in length with a GC content of 36.7%. The quadripartite structure of the cp genome has one LSC region of 90,585 bp, one SSC region of 18,959 bp, and a pair identical IR regions of 25,880 bp. A total of 117 genes were annotated including 80 protein-coding, 33 tRNA and four rRNA genes. Comparison of the L. hancei cp genome with the publicly available L. balansae genome using the program mVISTA (Mayor et al. Citation2000) under the LAGAN mode, indicated that the two were highly conserved, containing 80 protein-coding genes and four rRNA. However, L. hancei has 31 tRNA, while L. balansae contains 33 of these genes. The ML tree () fully supported a monophyletic Lithocarpus with L. hancei closely related to L. polystachyus Rehd. The cp genome of L. hancei will help us better construct a robust phylogeny of Lithocarpus and the Fagaceae in the future study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [http://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MW375417. The associated BioProject, SRA, and Biosample numbers are PRJNA684954, SRX9680931, and SAMN17073066, respectively.
Additional information
Funding
References
- Alzahrani DA, Yaradua SS, Albokhari EJ, Abba A. 2020. Complete chloroplast genome sequence of Barleria prionitis, comparative chloroplast genomics and phylogenetic relationships among Acanthoideae. BMC Genomics. 21(1):393.
- Birky CW, Maruyama T, Fuerst P. 1983. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics. 103(3):513–527.
- Cannon CH, Manos PS. 2003. Phylogeography of the Southeast Asian stone oaks (Lithocarpus). J Biogeogr. 30(2):211–226.
- Cao K-F. 2001. Morphology and growth of deciduous and evergreen broad-leaved saplings under different light conditions in a Chinese beech forest with dense bamboo undergrowth. Ecol Res. 16(3):509–517.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang CJ, Bruce Bartholo Z-Y. 1999. Flora of China. Vol. 4. Beijing: Science Press, Fagaceae; p. 333–369.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 16(11):1046–1047.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yang J, Vazquez L, Chen X, Li H, Zhang H, Liu Z, Zhao G. 2017. Development of chloroplast and nuclear DNA markers for Chinese oaks (Quercus Subgenus Quercus) and assessment of their utility as DNA barcodes. Front Plant Sci. 8:816.
- Yang Y, Zhou T, Duan D, Yang J, Feng L, Zhao G. 2016. Comparative analysis of the complete chloroplast genomes of five Quercus species. Front Plant Sci. 7:959.