Abstract
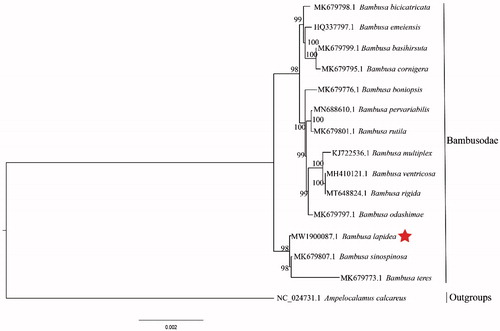
Bambusa lapidea is primarily distributed in Guangdong, Guangxi, Sichuan, Yunnan, and Hong Kong in China, occurring on plains, lower hills, and wetlands on both sides of rivers and adjacent to villages. Therefore, we sequenced and reported the complete chloroplast genome of B. lapidea for the first time. The complete chloroplast genome sequence of B. lapidea was generated by de novo assembly using whole-genome next generation sequencing. The genome was 139,525 bp in total length, including a large next-copy (LSC) region of 83,034 bp, a small single-copy (SSC) region of 12,893 bp, a pair of invert repeats (IR) regions of 21,799 bp. The plastid genome contained 127 genes including 83 protein-coding genes, 36 tRNA genes, and eight rRNA genes. Phylogenetic analysis based on 14 chloroplast genomes indicates that B. lapidea is closely related to B. arnhemica sinospinosa and B. teres in Bambusodae.
Bambusa lapidea (https:/wcsp.science.kew.org/qsearch.do) is known for its outstanding fibrous raw materials with a large-scale clump bamboo species of high development and consumption value (Shi et al. Citation2009). The chloroplasts (cp) genome has a maternal inheritance and conserved structure that has been used to examine the developmental and phylogenetic relationships of plants (Wang et al. Citation2018). In this study, we identified the complete cp genome sequence of B. lapidea using Illumina sequencing data to explain the species' phylogenetic position and to do further evolutionary research. B. lapidea leaves specimens were taken from Fujian Province, China (University of Fujian Agriculture and Forestry, Bamboo Garden, Fuzhou:119°14′16″E, 26°5′7″ N) and quickly dried with silica gel for DNA extraction. The specimens were stored in the Bamboo Research Institute, Fujian Agriculture and Forestry University (contact person name: Tianyou He & Email: [email protected]) under the voucher number 101302). The improved CTAB method (Doyle and Doyle Citation1987) was used to extract DNA of the fresh leaf sample and its quantification was validated using Agarose gel electrophoresis, and Nanodrop concentration @500 bp randomly interrupted by the Covaris ultrasonic breaker for library construction. Approximately, 2.0 GB of raw data were generated with 150 bp paired-end read lengths. The Illumina High-throughput sequencing platform (HiSeq2500) data were filtered by the script in the NOVOPlasty (Dierckxsens et al. Citation2017). The complete cp genome of Bambusa arnhemica (GeneBank accession: KJ870989) as a reference and plastid genome of B. lapidea both were assembled by GetOrganelle pipe-line (https://github.com/Kinggerm/GetOrganelle), it can get the plastid-like reads. The cp genome annotation was assembled based on the comparison by Geneious v 11.1.5 (Biomatters Ltd, Auckland, New Zealand) (Kearse et al. Citation2012; Jin et al. Citation2018).
The final complete cp genome sequence of B. Lapidea has been submitted to GenBank under the accession number MW190087. Raw reads were deposited in the GenBank Sequence Read Archive (SRA PRJNA687881).The complete cp genome sequence of B. Lapidea was a circular shape of 139,525 bp in length, consisting of four distinct regions, such as a large single-copy (LSC) region of 83,034 bp, a small single-copy (SSC) region of 12,893 bp, and a pair of inverted repeats (IRa and IRb) regions of 21,799 bp each. Furthermore, the complete cp genome consisted of 127 genes, including 83 protein coding genes, 36 tRNA genes, eight rRNA genes, and 38.90% of GC content. Phylogenetic analyses including B. lapidea, 13 other Bambusodae species and one outgroup of Arundinarodae,were performed using complete cp genomes. All of them were downloaded from NCBI GenBank. The sequences were aligned by MAFFT v7.307 (Katoh and Standley Citation2013), and the phylogenetic tree was constructed by RAxML (Stamatakis Citation2014). In this study, the phylogenetic tree revealed that B. lapidea is closely related to B. sinospinosa and B. teres (Bambusodae) as illustrated in ().
Acknowledgments
The authors wish to thank the anonymous reviewers who provided constructive reviews and critical input on this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GeneBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number MW190087. All high-throughput sequencing data files are available from the GenBank Sequence Read Archive (SRA) accession number: PRJNA687881.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Jin J-J, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv, p.256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment soft-ware version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Shi ZJ, Zhang JY, Wu CH, Qin YJ. 2009. Analysis of Bambusa lapidea material properties and evaluation of development and utilization value. J Anhui Agric Sci. 37(34):17180–17181.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang J, Li C, Yan C, Zhao X, Shan S. 2018. A comparative analysis of the complete chloroplast genome sequences of four peanut botanical varieties. PeerJ. 6:e5349.