Abstract
Polyalthopsis Chaowasku is a recently newly described genus in Annonaceae. Polyalthopsis verrucipes (C.Y.Wu ex P.T.Li) B.Xue & Y.H.Tan is distributed in Southern Yunnan, China. In this article, we report the complete chloroplast (cp) genome of Polyalthiopsis based on Illumina sequencing data. The whole cp genome of this species is 159,965 bp in length, consisting of two inverted repeat regions (IR, 25,974 bp each), one large single-copy region (LSC, 89,030 bp), and one small single-copy region (SSC, 18,987 bp). A total of 130 genes were annotated for the cp genome, including 83 protein-coding genes, 37 tRNAs, and 8 rRNAs. Phylogenetic analysis indicated that P. verrucipes was closely related to Meiogyne hainanensis.
The genus Polyalthiopsis Chaowasku is a recently published small genus in Annonaceae (Chaowasku, Damthongdee, et al. Citation2018; Xue et al. Citation2020). It includes one species in Vietnam and two species in China (Xue et al. Citation2020). It belongs to the tribe Miliuseae, and is sister to the genus Miliusa Lesch. ex A. DC. (Xue et al. Citation2020). The long-recognized sister relationship between Miliusa and Huberantha in previous phylogenetic studies (e.g. Chaowasku, Thomas, et al. Citation2014) has been redefined following the inclusion of Polyalthiopsis.
Polyalthiopsis verrucipes (C.Y.Wu ex P.T.Li) B.Xue & Y.H.Tan is distributed in Menghai, Yunnan, with restricted distribution and small populations. It was transferred from the genus Polyalthia Blume (Xue et al. Citation2020). This species is only represented by very few collections, and it is considered to be critically endangered as the habitat is severely destroyed (Xue et al. Citation2020).
In this study, we reported the complete chloroplast (cp) genome sequence of P. verrucipes, which represents the first plastid genome in the genus and the data would be helpful for the phylogenetic study of Polyalthiopsis and other closely related genera in the future.
The fresh leaves of Polyalthopsis verrucipes were collected from Menghai, Xishuangbanna, Yunnan province, China (21°46′22.5′′N, 100°02′58.5′′E). The total genomic DNA of the species was extracted using the modified CTAB method (Doyle and Doyle Citation1987). A voucher specimen (B. Xue & H.B. Ding XB311) was deposited in the herbarium of South China Botanical Garden, Chinese Academy of Sciences (IBSC) with the barcode number IBSC0860085. Library construction and sequencing were performed by BGI-Shenzhen (Shenzhen, China), using an Illumina HisSeq 2500 Sequencing System following the manufacturer’s instructions. The cp genome was assembled by the GetOrganelle (Jin et al. Citation2020). Using Meiogyne hainanensis (NC_043867.1) (recorded as Chieniodendron hainanense in Genbank) as a reference, the annotations were implemented using the online software Geseq (Tillich et al. Citation2017) with final manually correction. Finally, the complete sequences and annotations of P. verrucipes were submitted to GenBank with the accession number MW018366.
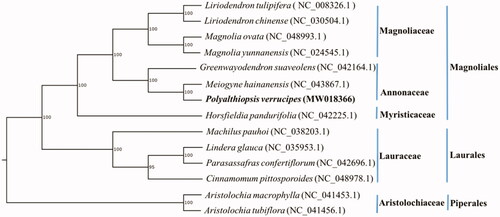
The complete cp genome of P. verrucipes is 159,965 bp in length, containing a large single-copy (LSC) region of 89,030 bp, a small single-copy (SSC) region of 18,987 bp, and two inverted repeat (IR) regions of 25,974 bp each. The whole cp genome has 130 genes in total, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. In addition, the overall GC content of the genome is 39.05%. In order to investigate the phylogenetic relationships of P. verrucipes with other species in Annonaceae and outgroup species, another two complete cp genomes from Annonaceae and 11 complete cp genomes from several closely related families Aristolochiaceae, Magnoliaceae, Myristicaceae, and Lauraceae were obtained from GenBank and aligned with P. verrucipes using MAFFT (Katoh and Standley Citation2013). A maximum likelihood analysis was performed by RAxML (Stamatakis Citation2014) under GTR + G model with 1000 bootstrap replicates. Aristolochia macrophylla (GenBank Accession NC_041453.1) and Aristolochia tubiflora (Genbank Accession NC_041456.1) were selected as outgroups. The phylogeny showed that P. verrucipes was closely related to genus Meiogyne in Annonaceae; the family Annonaceae is closely related to Magnoliaceae and Myristicaceae ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in GenBank (https://www.ncbi.nlm.nih.gov/) under the accession number MW018366.
Additional information
Funding
References
- Chaowasku T, Damthongdee A, Jongsook H, Nuraliev MS, Ngo DT, Le HT, Lithanatudom P, Osathanunkul M, Deroin T, Xue B, et al. 2018. Genus Huberantha (Annonaceae) revisited: erection of Polyalthiopsis, a new genus for H. floribunda, with a new combination H. luensis. Annales Botanici Fennici. 55(1–3):121–136.
- Chaowasku T, Thomas DC, van der Ham RWJM, Smets EF, J.B M, Chatrou LW. 2014. A plastid DNA phylogeny of tribe Miliuseae: insights into relationships and character evolution in one of the most recalcitrant major clades of Annonaceae. Am J Bot. 101(4):691–709.
- Doyle JJ, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Xue B, Ding HB, Yao G, Shao YY, Fan XJ, Tan YH. 2020. From Polyalthia to Polyalthiopsis (Annonaceae): transfer of species enlarges a previously monotypic genus. PhytoKeys. 148:71–91.