Abstract
Osmanthus fragrans is a well-known ornamental tree with high medicinal and edible values. In this study, the complete mitochondrial genome sequence of O. fragrans was assembled, annotated, and analyzed using phylogenomic methods. The complete mitochondrial genome of O. fragrans was 563,202 bp in length and displays an overall GC content of 44.58%. Sixteen chloroplast-derived segments with an average length of 1260 bp were identified. The complete mitochondrial genome contained 74 genes in total, including 44 protein-coding, three rRNA, and 27 tRNA genes, among which seven protein-coding and six tRNA genes were chloroplast-derived. Phylogenetic analysis showed that O. fragrans was closely related to Chionanthus rupicola within the Oleaceae. This study could provide genomic resources for a better understanding of O. fragrans and further studies on the evolution of Oleaceae.
Osmanthus fragrans Lour. (sweet osmanthus, Oleaceae) is a well-known evergreen ornamental tree that produces small white, yellow, or orange flowers with a sweet, rich fragrance (Xiang and Liu Citation2008). It is also edible and has medicinal values (Zhou et al. Citation2017). In previous phylogenetic studies, O. fragrans was clustered with Chionanthus retusus based on nuclear ribosomal DNA sequences (Besnard et al. Citation2009), which is inconsistent with the results based on plastid sequences (Besnard et al. Citation2009; Duan et al. Citation2019). In this study, the complete mitochondrial genome sequence of O. fragrans was assembled and characterized to reveal the evolutionary history of O. fragrans based on a mitochondrial phylogenomic analysis.
The fresh leaves were collected from a tree in the Shijiazhuang People’s Medical College (Hebei, China) (N 37.99°, E 114.45°). The voucher specimen (accession no. PMC160614) was deposited in the Shijiazhuang People’s Medical College (http://www.sjzrmyz.com/, Jie Guo, [email protected]). Genomic DNA was extracted using a modified CTAB method (Doyle and Doyle Citation1987). The extracted DNA was fragmented for Illumina library construction by Illumina TruSeq DNA sample prep kit and then sequenced on the HiSeq X Ten platform (Illumina Inc., San Diego, CA, USA). The complete mitochondrial genome sequence of O. fragrans was assembled with GetOrganelle v1.6.2e (Jin et al. Citation2020) and annotated with the OGAP pipeline using the default settings (https://github.com/zhangrengang/OGAP). The draft annotations were then adjusted manually with the assistance of BLAT (Kent Citation2002).
The complete mitochondrial genome of O. fragrans was assembled as a circular molecule 563,202 bp in length (GenBank accession no. MW645067), with an overall GC content of 44.58%. Sixteen chloroplast-derived segments were identified with lengths ranging from 190 to 3349 bp and an average length of 1260 bp. A total of 74 genes were annotated, including 44 protein-coding, three rRNA, and 27 tRNA genes. Among these, seven protein-coding and six tRNA genes were in the above-mentioned chloroplast-derived segments. The mitogenome size and composition of O. fragrans were similar to the four related taxa in the Oleaceae (), which contained 37–48 protein-coding, three rRNA, and 20–26 tRNA genes with a length ranging from 658,522 (Hesperelaea palmeri) to 848,451 bp (Ligustrum quihoui) and a GC content of 44.5–44.8% (Van de Paer et al. Citation2016, Citation2018).
Figure 1. Phylogenetic tree inferred by maximum-likelihood method based on mitochondrial protein-coding gene sequences of O. fragrans and other 12 species within the order Lamiales. Solanum lycopersicum was served as the outgroup. Numbers in the nodes are the bootstrap values from 1000 replicates. The branch of Ajuga reptans was truncated as it was too long.
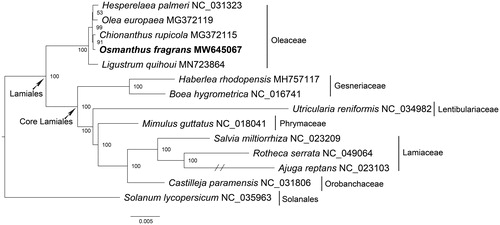
The phylogenetic analysis was performed with the mitochondrial genomes from 12 other species in the order Lamiales, designating Solanum lycopersicum (Solanales) as the outgroup. The 37 mitochondrial protein-coding genes of these species were aligned by using MAFFT v7.471 (Standley and Katoh Citation2013) and the alignment was trimmed with TrimAl (Capella-Gutierrez et al. Citation2009). The maximum likelihood phylogenetic tree was reconstructed by using the software IQ-TREE v1.6.5 (Nguyen et al. Citation2015) based on the best-fit model of TVM + F + R3 and 1000 bootstrap replicates. The phylogenetic tree showed that O. fragrans was closely related to Chionanthus rupicola and formed a monophyletic clade with four other species classified in the Oleaceae. This monophyletic clade was situated in a basal position of the Lamiales (). The phylogenetic relationships within the Lamiales were consistent with previous chloroplast-based studies (Duan et al. Citation2019; Zhu et al. Citation2020). In summary, this study extended our knowledge to O. fragrans and provided a reference for further research on the phylogeny and evolution of the family Oleaceae as well as the order Lamiales.
Disclosure statement
No potential conflict of interest was declared by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at http://www.ncbi.nlm.nih.gov/ under the accession no. MW645067. The associated BioProject, SRA, and BioSample numbers are PRJNA713943, SRR13948793, and SAMN18275439, respectively.
References
- Besnard G, Rubio de Casas R, Christin PA, Vargas P. 2009. Phylogenetics of Olea (Oleaceae) based on plastid and nuclear ribosomal DNA sequences: tertiary climatic shifts and lineage differentiation times. Ann Bot. 104(1):143–160.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantity of fresh leaf material. Phytochem Bull. 119:11–15.
- Duan Y, Li Y, Zhang C, Wang X, Li M. 2019. The complete chloroplast genome of sweet olive (Osmanthus fragrans Lour.). Mitochondrial DNA Part B. 4(1):1063–1064.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kent WJ. 2002. BLAT-the BLAST-like alignment tool . Genome Res. 12(4):656–664.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Standley DM, Katoh K. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Van de Paer C, Hong-Wa C, Jeziorski C, Besnard G. 2016. Mitogenomics of Hesperelaea, an extinct genus of Oleaceae. Gene. 594(2):197–202.
- Van de Paer C, Bouchez O, Besnard G. 2018. Prospects on the evolutionary mitogenomics of plants: a case study on the olive family (Oleaceae). Mol Ecol Resour. 18(3):407–423.
- Xiang Q, Liu Y. 2008. An illustrated monograph of the sweet osmanthus variety in China. Hangzhou: Zhejiang Science and Technology Press.
- Zhou F, Zhao Y, Li M, Xu T, Zhang L, Lu B, Wu X, Ge Z. 2017. Degradation of phenylethanoid glycosides in Osmanthus fragrans Lour. flowers and its effect on anti-hypoxia activity. Sci Rep. 7(1):10068.
- Zhu X, Yang K, Xiong Z, Li X. 2020. The complete chloroplast genome of Silvianthus bracteatus (Carlemanniaceae) and phylogenic analysis of Lamiales. Mitochondrial DNA Part B. 5(2):1132–1133.
