Abstract
We report the complete mitochondrial genome sequence of the endangered Eurasian otter, Lutra lutra. The complete mitochondrial genome is 16,537 bp in length and contains 13 protein-coding genes, 22 transfer RNA, two ribosomal RNA, and one control region. The mitogenome is A + T rich, with a composition of 32.2% A, 27.5% C, 14.5% G, and 25.8% T. Phylogenetic analysis based on 13 protein-coding mitochondrial genes of Mustelidae supports the conventional systematic treatment with eight subfamilies. Lutra lutra is closely related to Lutra sumatrana and the subfamily Lutrinae was closely grouped with the Ictonychinae. This study provides genetic and taxonomic information for future studies of Eurasian otters and the Mustelidae.
The Eurasian otter, Lutra lutra (Linnaeus, 1758) (Carnivora, Mustelidae, and Lutrinae) is a cosmopolitan species widely distributed from Western Europe to north-eastern Siberia and the Korean Peninsula (Jo et al. Citation2018). The status of Eurasian otters has been regarded as “near threatened” by the Red List of the International Union for Conservation of Nature (IUCN). Also, the Conservation on International Trade in Endangered Species (CITES) has listed the species in Appendix I (Roos et al. Citation2015). European populations of Lutra lutra have been declining and some local populations have been extirpated (Ferrari et al. Citation2017). Despite the global decline and social interests, only a limited number of genetic studies on the Eurasian otter have been published (Jo et al. Citation2012, Citation2017). In particular, few genomic studies have been performed (NIBR Citation2019).
Currently, the Mustelidae including the otter subfamily, Lutrinae, is subdivided into eight subfamilies based on the limited number of genetic markers and morpho-anatomical studies of fossils (Koepfli et al. Citation2008; Law et al. Citation2018). Unfortunately, the limited numbers of genetic markers or small marker lengths can sometimes fail to fully resolve the true evolutionary relationships among populations (Koepfli and Wayne Citation2003; Yu et al. Citation2008). To clarify the phylogenetic analysis of the subfamily and family, more molecular markers are required (Morton and Telmer Citation2014).
Here we sequenced the complete mitochondrial genome for Lutra lutra and analyzed the phylogenetic history of members classified to the subfamilies of Mustelidae. The voucher specimen was a road-killed Eurasian otter from the Daejeon metropolitan area in South Korea (36°21′35.13″N 127°22′46.22″E), and the study skin and skull with extracted DNA are stored at the Daegu University in South Korea (Voucher No. DUMM0035, the animal specimen storage room in the department of biology education [https://bioedu.daegu.ac.kr/] curator: Prof. Y. S. Jo [[email protected]]). DNA was extracted from muscle tissue using a QIAGEN blood and tissue kit (Qiagen, Valencia, CA, USA), and genomic DNA was sequenced using the HiseqX platform (Illumina, San Diego, CA, USA). The assembly was performed de novo using the low coverage whole genome shotgun sequencing (dnaLCW) method from PHYGEN (Seongnam, Korea). We employed the GeSeq program (https://chlorobox.mpimp-golm.mpg.de/geseq-app.html, Tillich et al. Citation2017) for the annotation.
The complete mitochondrial genome of L. lutra is 16,537 bp in length (MW573979) and contains 13 protein-coding genes (ND1-ND6; COXI-3; ATP6 and ATP8; CYT B), 22 transfer RNA, two ribosomal RNA, and one control region (D-loop). The base composition of the mitochondrial genome of Eurasian otters from Korea is 32.2% A, 27.5% C, 14.5% G, and 25.8% T with the total A-T content (58%) higher than G–C content (42%). The 12S rRNA and 16 s rRNA are 964 and 1568 bp, respectively. The 12S rRNA gene is located between RNAPhe and tRNAVal. and the 16S rRNA gene is positioned between tRNAVal and tRNALeu. The ND and eight tRNAs are encoded in the reverse strand, while the remainder is on the forward strand. There are 10 PCGs initiate with the ATG as a start codon, while ND2 initiates with ATC, and ND3 and ND5 begin with ATA. Incomplete stop codons are found in ND2 and COX3 (T-), and ND1 (TA-). For Cytb, AGA terminates the gene and the other nine genes terminate with TAA.
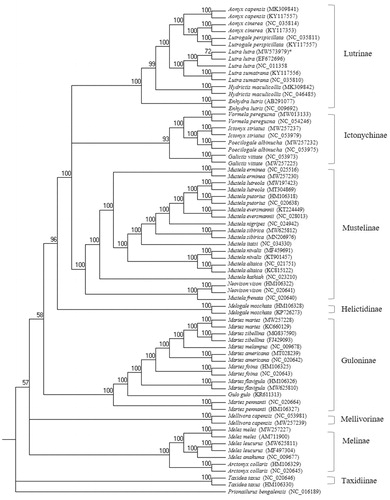
For the phylogenetic analysis, 13 protein-coding genes (PCGs) representing eight subfamilies were downloaded from GenBank, including five genera of Lutrinae. As the outgroup, the Leopard cat, Prionailurus bengalensis (Kerr, 1792) (Carnivora, Felidae) (KP246843) was designated. Seventy-one individuals mitochondrial PCG sequences from 39 species were aligned with MAFFT v.7040 using default settings (Katoh et al. Citation2002; Katoh and Standley Citation2013). We employed RAxML v.8.2.11 (Stamatakis Citation2014) for reconstructing the phylogenetic tree with 1000 bootstrap replicates and the default general time-reversible GTR + GAMMA model. The model supports the conventional eight monophyletic subfamilies in Mustelidae (). The Korean population of the otter forms a sister group to Lutra sumatrana (bootstrap value, 100). The Asian small-clawed otter, Aonyx cinerea, was grouped with the smooth-coated otter, Lutrigale perspicillata, rather than the Asian clawless otter, Aonyx capenis. Although the previous studies reported that the Mustelinae is the closest subfamily of the Lutrinae (Moretti et al. Citation2017), these results show that the Ictonychinae and Lutrinae were unresolved in a clade separate from the Mustelinae, Helictidinae, and Guloinae. These results based on mitochondrial genomic markers generally coincided with the previous studies (Koepfli et al. Citation2008; Law et al. Citation2018). The Korean L. lutra from Daejeon differed from L. lutra from other specimens from Korea, GenBank accession number NC_011358 (exact location is unknown, expect Korea; Jang et al. Citation2009 ) by 25 SNPs and one indel (9 bp in the length), from GenBank accession number EF672696 (exact location is unknown, expect Korea; Ki et al. Citation2010) by 29 SNPs and two indels (12 and 15 bp in the length). The genomic resources of the Korean L. lutra obtained from this study provide an additional reference for genetic studies as well as conservation and management for the Eurasian otters.
Figure 1. The phylogenetic relationship of the Lutra luta and other Mustelidae species based on 13 PCGs sequences using RAxML program with Prionailurus bengalensis as an outgroup. The numbers on the branches indicate bootstrap value. *Indicates the Korean otter Lutra lutra used in this study with the mitogenome accession no. MW573979.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov, accession no. MW573979. The associated BioProject, SRA, and BioSample numbers are PRJNA725629, SRR14478347, and SAMN18897176, respectively. The data that support the findings of this study are also openly available in Mendeley Data at http://dx.doi.org/10.17632/74bpx3kfx3.1.
References
- Ferrari C, D'Alfonso M, Moris V, Battisti A, Giuliano D. 2017. Diet in a free ranging individual of Lutra lutra in Valsavarenche. J Mt Ecol. 10:7–12.
- Jang K, Ryu S, Hwang U. 2009. Mitochondrial genome of the Eurasian otter Lutra lutra (Mammalia, Carnivora, Mustelidae). Genes & Genomics 31(1):19–27.
- Jo Y, Baccus J, Koprowski J. 2018. Mammals of Korea. Incheon: NIBR.
- Jo Y, Won C, Fritts S, Wallace M, Baccus J. 2017. Distribution and habitat models of the Eurasian otter, Lutra lutra, in South Korea. J. Mamm. 98(4):1105–1117.
- Jo Y, Won C, Jung J. 2012. Testing microsatellite loci and preliminary genetic study for Eurasian otter in South Korea. J. Species Res. 1(2):240–248.
- Katoh K, Misawa K, Kuma KI, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Ki J, Hwang D, Park T, Hang S, Lee J. 2010. A comparative analysis of the complete mitochondrial genome of the Eurasian otter Lutra lutra (Carnivora; Mustelidae). Mol. Biol. Rep. 37:1943–1955.
- Koepfli KP, Deere KA, Slater GJ, Begg C, Begg K, Grassman L, Lucherini M, Veron G, Wayne RK. 2008. Multigene phylogeny of the Mustelidae: resolving relationships, tempo and biogeographic history of a mammalian adaptive radiation. BMC Biol. 6(1):10.
- Koepfli KP, Wayne RK. 2003. Type I STS markers are more informative than cytochrome b in phylogenetic reconstruction of the Mustelidae (Mammalia: Carnivora). Syst Biol. 52(5):571–593.
- Law CJ, Slater GJ, Mehta RS. 2018. Lineage diversity and size disparity in Musteloidea: testing patterns of adaptive radiation using molecular and fossil-based methods. Syst Biol. 67(1):127–144.
- Moretti B, Al-Sheikhly OF, Guerrini M, Theng M, Gupta BK, Haba MK, Khan WA, Khan AA, Barbanera F. 2017. Phylogeography of the smooth-coated otter (Lutrogale perspicillata): distinct evolutionary lineages and hybridization with the Asian small-clawed otter (Aonyx cinereus). Sci Rep. 7(1):41611–41613.
- Morton CM, Telmer C. 2014. New subfamily classification for the Rutaceae. Ann Missouri Bot Gard. 99(4):620–641.
- NIBR. 2019. Genetic diversity of animal resources IV-2. Incheon: National Institute of Biological Resources.
- Roos A, Loy A, de Silva P, Hajkova P, Zemanová B. 2015. Lutra lutra. The IUCN Red list of threatended species 2015: e. T12419A21935287.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yu L, Liu J, Luan PT, Lee H, Lee M, Min MS, Ryder OA, Chemnick L, Davis H, Zhang YP. 2008. New insights into the evolution of intronic sequences of the beta-fibrinogen gene and their application in reconstructing mustelid phylogeny. Zool Sci. 25(6):662–672.
