Abstract
Colias fieldii is a common seen diurnal butterflies in the fields and widely distributed in many provinces of China. In this study, we sequenced and analyzed the complete mitochondrial genome (mitogenome) of C. fieldii. This mitogenome was 15,150 bp long and encoded 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA unit genes (rRNAs). The overall base composition of the mitogenome was estimated to be A 39.8%, T 41.2%, C 11.4% and G 7.6%, with a high A + T content of 81.0%. Except for cox1 started with CGA, all other PCGs started with the standard ATN codons (seven ATG, four ATT and one ATC). Most of the PCGs terminated with the stop codon TAA or TAG, whereas cox1, cox2, nad5 and nad4 end with the incomplete codon T––. Phylogenetic analysis showed that C. fieldii is indeed the sister species of Colias erate with a high support value. All seven Coliadinae species formed one clade and was sister to Pierinae butterflies. Within Coliadinae, the relationships (Eurema + (Gonepteryx + (Catopsilia + Colias))) were highly supported.
The genus Colias contains 84 species and almost 120 subspecies. They are adapted to different habitats and can be divided into mountain species, lowland species, and species inhabiting medium elevations in generally speaking (Stella et al. Citation2018). The wings of Colias butterflies are red, orange, yellow, or greenish, often with a black rim on the outer side. Colias butterflies occur throughout the Holarctic, including the arctic regions, and sexual dimorphism or polymorphism is expressed in this genus (Srygley and Kingsolver Citation1998). Colias fieldii Ménétriès, 1855 is a butterfly which has bright orange color wings, and it can damage soybean, alfalfa and trefoil. Mitogenome can be utilized in research on population genetic structure, taxonomic resolution, phylogeography and phylogeny. For taxonomic resolution and further study on population genetic structure of C. fieldii, we sequenced the complete mitogenome of C. fieldii and analyzed the phylogenetic relationships of Pieridae based on mitogenome data.
Specimens of C. fieldii were collected from Taihe County, Jiangxi Province, China (26°49′N, 114°52′E, August 2019) and were stored in Institute of Plant Protection of Qinghai Academy of Agricultural and Forestry Sciences (please contact Dr. Yunxiang Liu, email: [email protected]) under the voucher number QHAF-ECF02. Total genomic DNA was extracted from muscle tissues of the thorax using DNeasy DNA Extraction kit (Qiagen, Hilden, Germany). A pair-end sequence library was constructed and sequenced using Illumina HiSeq 2500 platform (Illumina, San Diego, CA), with 150 bp pair-end sequencing method. A total of 26.6 million reads were generated and had been deposited in the NCBI Sequence Read Archive (SRA) with accession number SRR14560146. With the mitochondrial genome of Colias erate (KP715146) employed as reference, raw reads were assembled using MITObim v 1.7 (Hahn et al., Citation2013). By comparison with the homologous sequences of other Pieridae species from GenBank, the mitogenome of C. fieldii was annotated using software GENEIOUS R11 (Biomatters Ltd., Auckland, New Zealand).
The complete mitogenome of C. fieldii is 15,150 bp in length (GenBank accession no. MT371042), containing the typical set of 13 protein-coding, two rRNA and 22 tRNA genes, and one non-coding AT-rich region. The overall base composition of the mitogenome was estimated to be A 39.8%, T 41.2%, C 11.4% and G 7.6%, with a high A + T content of 81.0%. Except for cox1 started with CGA, all other PCGs started with the standard ATN codons (seven ATG, four ATT and one ATC). Most of the PCGs terminated with the stop codon TAA or TAG, whereas cox1, cox2, nad5 and nad4 end with the incomplete codon T––. Gene order was conserved and identical to most other previously sequenced Pieridae butterflies (Park et al. Citation2012; Hao et al. Citation2013; Cao et al. Citation2016; Fang et al. Citation2016; Nie et al. Citation2018). The 22 tRNA genes vary from 60 bp (trnS1) to 70 bp (trnK). Two rRNA genes (rrnL and rrnS) locate at trnL1/trnV and trnV/control region, respectively. The lengths of rrnL and rrnS in M. calida are 1327 and 770 bp, with the AT contents of 85.0% and 84.9%, respectively.
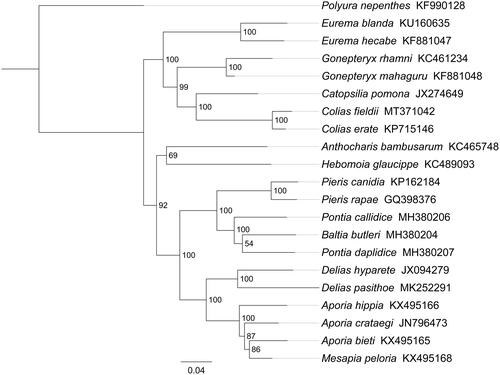
Phylogenetic analysis was performed based on the nucleotide sequences of 13 PCGs from 21 Papilionoidea species. Phylogenetic tree was constructed through raxmlGUI 1.5 (Silvestro and Michalak Citation2012). Results showed that C. fieldii is indeed the sister species of Colias erate with a high support value (BS = 100) (). All seven Coliadinae species formed one clade and was sister to Pierinae butterflies. Within Coliadinae, the relationships (Eurema + (Gonepteryx + (Catopsilia + Colias))) were highly supported, and similar results were found in the previous work (Ding and Zhang Citation2017; Zhou et al. Citation2020). In conclusion, the mitogenome of C. fieldii is sequenced in this study and can provide essential DNA molecular data for further phylogenetic and evolutionary analysis of Pieridae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MT371042. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA730322, SRR14560146, and SAMN19228343 respectively.
Additional information
Funding
References
- Cao Y, Hao JS, Sun X, Zheng B, Yang Q. 2016. Molecular phylogenetic and dating analysis of pierid butterfly species using complete mitochondrial genomes. Genet Mol Res. 15(4).
- Ding C, Zhang Y. 2017. Phylogenetic relationships of Pieridae (Lepidoptera: Papilionoidea) in China based on seven gene fragments. Entomol Sci. 20(1):15–23.
- Fang J, Wu Y, Wang H, Sun Z, Han D, Zhang B. 2016. The complete nucleotide sequence of the mitochondrial genome of the Oriental Pieris, Pieris canidia (Lepidoptera: Pieridae). Mitochondrial DNA Part A. 27(6):4088–4089.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Hao J, Wang Y, Sun X, Zhang L, Hao J, Yang Q. 2013. The complete mitochondrial genome of Hebomoia glaucippe (Lepidoptera: Pieridae). Mitochondrial DNA. 24:668–670.
- Nie L, Wang Y, Huang D, Tao R, Su C, Hao J, Zhu C. 2018. Mitochondrial genomes of four pierid butterfly species (Lepidoptera: Pieridae) with assessments about Pieridae phylogeny upon multiple mitogenomic datasets. Zool Syst. 43:387–409.
- Park JS, Cho Y, Kim MJ, Nam SH, Kim I. 2012. Description of complete mitochondrial genome of the black-veined white, Aporia crataegi (Lepidoptera: Papilionoidea), and comparison to papilionoid species. J Asia-Pac Entomol. 15(3):331–341.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Srygley RB, Kingsolver JG. 1998. Red-wing blackbird reproductive behaviour and the palatability, flight performance, and morphology of temperate pierid butterflies (Colias, Pieris, and Pontia). Biol J Linn Soc. 64(1):41–55.
- Stella D, Falt Nek Fric Z, Rindoš M, Kleisner K, Pech Ček P. 2018. Distribution of ultraviolet ornaments in Colias butterflies (Lepidoptera: Pieridae). Environ Entomol. 47(5):1344–1354.
- Zhou Y, Zhang C, Wang S, Liu Y, Wang N, Liang B. 2020. A mitogenomic phylogeny of pierid butterflies and complete mitochondrial genome of the yellow tip Anthocharis scolymus (Lepidoptera: Pieridae). Mitochondrial DNA Part B. 5(3):2587–2589.