Abstract
Rhamnus crenata Siebold & Zuccarini is deciduous shrub or small tree plant that widely distributed in temperate and tropical regions of East to Southeast Asia. It has a chloroplast genome structure similar to that of other species of Rhamnus. It is 160,454 bp in size, including a large single-copy (LSC) region of 88,884 bp, a small single-copy (SSC) region of 18,734 bp and a pair of inverted repeats (IRs) of 26,418 bp. A total of 122 genes were annotated, including 80 protein-coding genes, 38 transfer RNA (tRNA) genes, and 4 rRNA genes. The overall GC content is 30.71%. Phylogenetic analysis shows that R. crenata is clustered with R. taquetii and R. globosa.
Rhamnus crenata Siebold & Zuccarini (Rhamnaceae) is a deciduous shrub or small tree plant that widely distributed in mountain forests or thickets in temperate and tropical regions of East to Southeast Asia (Chen and Schirarend Citation2007). It has been used for a variety of ornamental purposes including hedge, privacy screen, windbreak and backdrop for perennial plantings. In this study, we characterized the complete chloroplast genome sequence of R. crenata (DDBJ accession number: LC635131) which would be helpful for phylogeny genetics research of Rhamnus and Rhamnaceae.
Fresh leaves of R. crenata without disease were collected from Ningbo, Zhejiang Province, China(N 29°44′14ʺ, E 121°06′45ʺ, 590 m)and dried immediately using silica gel in the field. The voucher specimen was deposited in the Herbarium of Southwest Forestry University (SWFC) (http://bbg.swfu.edu.cn, Bin Tian, [email protected]) under collection numbers of WJF2019003. We used DNeasyTM Tissue Kit (Qiagen, Hilden, Germany) to isolate total genomic DNA from dried leaves, and DNA samples were properly stored at Key Laboratory of Biodiversity Conservation in Southwest China, State Forestry Administration, Southwest Forestry University. The genomic paired-end (PE150) sequencing was performed on an Illumina Hiseq 2000 instrument (Illumina, Inc., San Diego, CA, USA). The chloroplast (cp) genome was assembled using the program Getorganelle (Jin et al. Citation2020). Annotation was performed using PGA (Qu et al. Citation2019) and visualized with OGDRAW (Greiner et al. Citation2019), coupled with manual correction for start and stop codons of protein-coding genes.
The cp genome of R. crenata resulted in a typical circular quadripartite structure of 160,454 bp length, with a LSC region of 88,884 bp, a SSC of 18,734 and two inverted repeats (IRa and IRb) each with a length of 26,418 bp. A total of 126 genes were annotated, including 84 protein-coding genes, 34 transfer RNA (tRNA) genes, and 8 rRNA genes. The overall GC content is 30.71%.
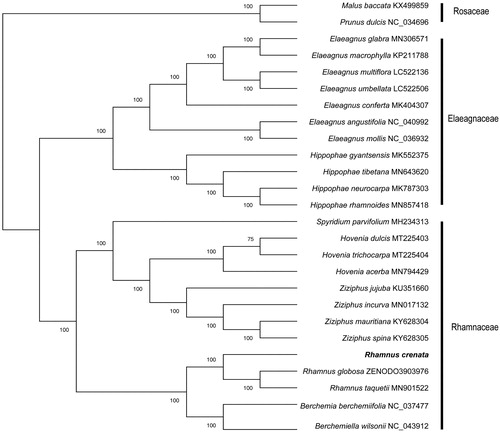
To verify the phylogenetic position of the newly obtained plastome of R. crenata and further clarify the evolutionary relationships within family Rhamnaceae, phylogenetic analyses based on total 26 complete cp genomes performed. After cp genomes was aligned via mafft (v.7.407) software (Katoh and Toh Citation2010) and removed gaps with Gblocks (Castresana Citation2000), Maximum Likelihood (ML) phylogenetic tree was constructed in the iqtree v.1.6.12 software (Nguyen et al. Citation2015) with bootstrap 1000 replicates and GTR + I model. The ML tree shows very clear results and most of the nodes had 100% bootstrap that Rhamnus crenata was belonged to Rhamnus and clustered with R. taquetii and R. globosa. (). Our findings provide a foundation for further investigation of chloroplast genome evolution in Rhamnaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in The DNA Data Bank of Japan (DDBJ; http://www.ddbj.nig.ac.jp) under the accession no. LC635131. The associated BioProject, SRA and Bio-Sample numbers are PRJDB11553, DRR288488, and SAMD00298364, respectively.
References
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Chen Y, Schirarend C. 2007. Rhamnaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, vol 12, Hippocastanaceae through Theaceae. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press. P141.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241–271.
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26(15):1899–1900.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.